Figure 7.
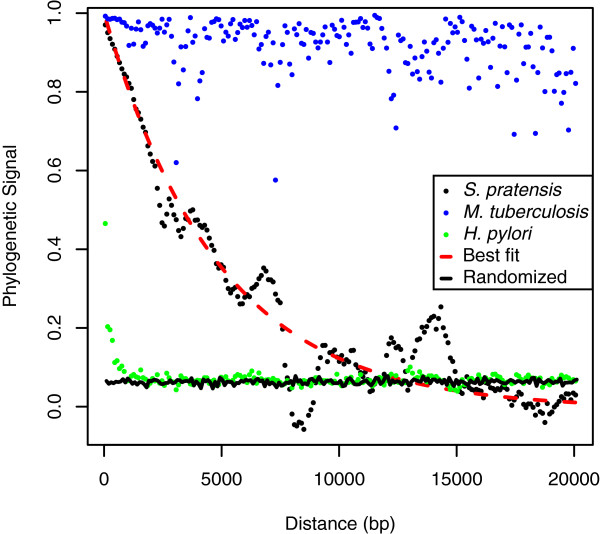
Recombination erases the intraspecies phylogenetic signal in S. pratensis. The proportion of incompatible sites along the five S. pratensis chromosomes is a function of chromosome distance. Phylogenetic signal is defined by a value of 1 indicating an absence of incompatible sites (pure vertical pattern of inheritance) and a value of 0 indicating that incompatible sites are randomly distributed by recombination. The solid black line indicates a random equilibrium SNP distribution generated from shuffling all SNPs among the S. pratensis genomes. Results are shown for the analysis of five S. pratensis genomes (black circles), as well as five Mycobacterium tuberculosis genomes which are known to represent a highly clonal species (blue circles) and five Helicobacter pylori genomes which are known to represent a highly recombining population (green circles). An exponential decay is fitted to the S. pratensis decay in phylogenetic signal due to distance between sites (dashed red line).
