Abstract
The diversity and success of teleost fishes (Actinopterygii) has been attributed to three successive rounds of whole-genome duplication (WGD). WGDs provide a source of raw genetic material for evolutionary forces to act upon, resulting in the divergence of genes with altered or novel functions. The retention of multiple gene pairs (paralogs) in teleosts provides a unique opportunity to study how genes diversify and evolve after a WGD. This study examines the hypothesis that vitamin D receptor (VDR) paralogs (VDRα and VDRβ) from two distantly related teleost orders have undergone functional divergence subsequent to the teleost-specific WGD. VDRα and VDRβ paralogs were cloned from the Japanese medaka (Beloniformes) and the zebrafish (Cypriniformes). Initial transactivation studies using 1α, 25-dihydroxyvitamin D3 revealed that although VDRα and VDRβ maintain similar ligand potency, the maximum efficacy of VDRβ was significantly attenuated compared with VDRα in both species. Subsequent analyses revealed that VDRα and VDRβ maintain highly similar ligand affinities; however, VDRα demonstrated preferential DNA binding compared with VDRβ. Protein-protein interactions between the VDR paralogs and essential nuclear receptor coactivators were investigated using transactivation and mammalian two-hybrid assays. Our results imply that functional differences between VDRα and VDRβ occurred early in teleost evolution because they are conserved between distantly related species. Our results further suggest that the observed differences may be associated with differential protein-protein interactions between the VDR paralogs and coactivators. We speculate that the observed functional differences are due to subtle ligand-induced conformational differences between the two paralogs, leading to divergent downstream functions.
Whole-genome duplication (WGD) events are proposed to be a significant force driving vertebrate evolution by providing a novel source of raw genetic material that can be subjected to evolutionary change (1–4). The evolution and divergence of new genes after a WGD may permit the development of more complex gene interactions and networks, leading to novel evolutionary innovations, speciation, and increased adaptability (3–7). Two WGD events (1R and 2R) occurred early in vertebrate evolution (Supplemental Figure 1) (1). A third WGD (3R) occurred in the stem lineage of teleost fish, a division within the vertebrate class Actinopterygii (ray finned fishes) (8, 9) (Supplemental Figure 1). It has been proposed that the 3R event has enabled significant diversification in teleosts (8, 10). For example, teleost fish compose approximately half of all species of vertebrates, whereas less than 50 species are found in pre-3R orders of Actinopterygii (11).
After a WGD, an estimated 50%–90% of gene duplicates become nonfunctional and are lost (12, 13). However, genes involved in signaling and regulatory pathways, such as the nuclear receptor (NR) superfamily, have been preferentially retained in duplicate to maintain pathway integrity (6). NRs are conditional transcription factors that bind to lipophilic signaling molecules, resulting in systematic control and expression of target genes. Such control facilitates cellular responses to endogenous and exogenous signals through the coordination of complex transcriptional processes (14). NRs play an essential role in many physiological and endocrine-mediated processes, including metabolism, reproduction, embryonic development, and cellular proliferation and differentiation. In teleosts, orthologs for all mammalian NR subclasses have been identified (15–17). In fact, teleosts maintain a larger complement of NRs than mammals, likely due to the global retention of NRs subsequent to the 3R event. For example, 48 NRs have been identified in humans (18), compared with 68–70 identified in pufferfishes (Takifugu rubripes and Tetraodon nigroviridis) (15, 16, 19), 70 in zebrafish (Danio rerio) (17), and 71 in the Japanese medaka (Oryzias latipes) (20). Because NRs regulate numerous processes essential to life, NR duplication and divergence may have contributed to the evolutionary success of teleosts. However, the functional role of duplicate NRs remains unresolved in these species.
The vitamin D receptor (VDR; NR1I1) is a member of the NR1I subfamily of nuclear receptors (14). Conventional theory speculates that the vitamin D endocrine system originated in terrestrial animals as a way to control calcium homeostasis in a calcium-limited environment (21). However, this theory has been challenged due to the identification of functional VDRs in numerous aquatic vertebrates including amphibians (21), numerous species of fish (15–17, 22–24), and the sea lamprey (25). The fact that aquatic vertebrates live in an environment that serves as an abundant calcium source suggests that VDR evolved long before the need for strict hormonal control of calcium.
In agreement with the 3R event, two distinct VDR genes (VDRα and VDRβ) have been identified in multiple 3R teleost species (15–17, 22, 23). Evidence indicating that these duplicates are indeed paralogs, and not isoforms, include the observation that VDRα and VDRβ paralogs are shared with multiple species across large evolutionary distances, the observation that they appear to have originated at the same time, and that the paralogs are found within distinct chromosomal loci yet maintain conserved synteny (8, 26). Homology comparisons have revealed that the VDR paralogs maintain a high degree of sequence identity to tetrapod VDRs, although are most similar to VDR paralogs from other teleosts (22). Tissue expression studies revealed both VDR paralogs are expressed in a wide variety of tissues similar to mammalian VDRs; however, VDRα and VDRβ exhibited differential spatial or quantitative tissue expression patterns in each species (15, 22, 27). Although VDR paralogs are highly conserved, studies have identified differential paralog functions, including differences in ligand sensitivities and target gene repertoire, which suggests a divergence of VDR function between paralogs at the molecular level (22, 27). The identification of divergent NR functions between VDRα and VDRβ may reveal important clues driving VDR evolution.
In this study, we build upon an initial observation that VDRα and VDRβ paralogs from the Japanese medaka exhibit differential sensitivities to 1, 25-dihydroxyvitamin D3 (1, 25D3) (22). Here we examine multiple steps in the VDR activation pathway to test the hypothesis that VDR paralogs from two distantly related phyla have undergone functional divergence subsequent to the 3R event.
Materials and Methods
DNA constructs
Medaka and zebrafish VDRα and VDRβ sequences were identified through screening the medaka and zebrafish genomes within Ensemble (28). Primers were designed in Primer3 (29) (see Table 1). Full-length VDRα and VDRβ were amplified and cloned into the TA cloning vector pCR2.1 (Life Technologies) and further subcloned into the pSG5 and pVP16 expression vectors as described elsewhere (22). Full-length VDR cDNAs were additionally subcloned into the pET32a expression vector (Novagen) for bacterial recombinant protein production. All constructs consisted of the complete VDR open reading frame including the internal start and stop codons except for pET32a in which the stop codons were removed for the inclusion of 3′ His tags. Sequence data for zebrafish VDRα (KJ925048) and VDRβ (KJ925049) have been submitted to GenBank. Medaka VDRα (EU403115) and VDRβ (EU403116) were submitted previously (22).
Table 1.
VDR Primer Sequences
| Name | Vector | R.S.a | F/R | Sequence (5′–3′) |
|---|---|---|---|---|
| Zebrafish VDRα | pSG5/pVP16 | EcoRI | F | GCG AAT TCG CCA TGC TTA CGG AAA ATA GTG CC |
| BamHI | R | ATG GAT CCA AAC TAG GAC ACC TCA CTC C | ||
| pET32a | SalI | F | GAT AGT CGA CTG ATG GAT CTG ATG GCC GTG | |
| NotI | R | GAT TGC GGC CGC ACT GGA CAC CTC ACT CC | ||
| Zebrafish VDRβ | pSG5/pVP16 |
EcoRI BamHI |
F | GCG AAT TCA TGG AGT CAG CTG TCA GTA C |
| R | ATG GAT CCA GAA AAC TAG GTG ACC TGC C | |||
| pET32a | SalI | F | GAT AGT CGA CTG ATG GAG TCA GCT GTC AGT | |
| NotI | R | GAT TGC GGC CGC ACT GGT GAC CTG CCC GCC | ||
| Medaka VDRα | pSG5 | BglII | F | GAA GAT CTA TGG AGT CCA TTA CCG TGA C |
| BglII | R | CGA GAT CTC TAT GAC ACC TCG CTG CCG A | ||
| pVP16 | SalI | F | CTC GTC GAC TTA TGG AGT CCA TTA CG TG | |
| HindIII | R | GCT AAG CTT CTC TAT GAC ACC TCG CTG CC | ||
| pET32a | SalI | F | GAT AGT CGA CTG ATG GAG TCC ATT ACC GTG | |
| NotI | R | GAT TGC GGC CGC ACT TGA CAC CTC GCT GCC | ||
| Medaka VDRβ | pSG5 | EcoRI | F | GCG AAT TCA TGG AGG CCA CTG TTG TGA G |
| BglII | R | CGA GAT CTC TAG GAG ACC TCG CTG CCA A | ||
| pVP16 | EcoRI | F | CCA GAA TTC ATG GAG GCC ACT GTT GTG | |
| HindIII | R | ATT AAG CTT CTC TAG GAG ACC TCG CTG CC | ||
| pET32a | SalI | F | GAT AGT CGA CTG ATG GAG GCC ACT GTT GTG | |
| NotI | R | GAT TGC GGC CGC ACT GGA GAC CTC GCT GCC |
Abbreviations: F, forward; R, reverse.
Restriction sites (R.S.) are indicated in bold lettering.
All coregulator expression constructs for transient transactivation studies (human pSG5-SRC1, pSG5-GRIP1, pCDNA-ACTR, pCDNA-RXRWT, pCDNA-RXRAF2) and Gal4 bait constructs for mammalian two-hybrid studies (human pM-SRC1241–386, pM-GRIP1479–767, pM-ATCR392–1005, pM-RXRWT, pM-RXRAF2) were a gift from Dr Donald McDonnell (Duke University, Durham, North Carolina). The RXRAF2 mutant is a truncated retinoid X receptor (RXR) that lacks H12, which contains the activation function-2 (AF2) region, because of an internal stop codon (D444Stop). The luciferase reporters XREM-Luc, 5XGal4-TATA-Luc, and pRL-CMV were obtained as previously reported (22).
Transient transactivation assays
Transactivation studies were conducted in HepG2 cells. This cell line exhibits fast growth kinetics and little to no background VDR activity and maintains active and diffusive transport mechanisms for the uptake of both polar and nonpolar ligands and high transfection efficiencies. HepG2 cells were maintained as described previously (22). Cells were seeded in 96-well plates (2.5 × 104 cells/well) and transfected 24 hours later using Lipofectamine 2000 (Life Technologies) following the manufacturer's recommendations. Cells were transfected with 89.7 ng of medaka or zebrafish pSG5-VDRα or pSG5-VDRβ, 19.2 ng of the XREM-Luc reporter (30), and 4.5 ng of pRL-CMV as an internal luciferase control.
Coregulator studies included 18.3 ng of an expression vector containing the complete open reading frame of the coregulator of interest (pCDNA-RXRWT or RXRAF2 and/or pSG5-SRC1, pSG5-GRIP1, or pCDNA-ACTR). Media were replaced the following day with complete MEM containing 120 nM 1, 25D3 (EMD Millipore) in ethanol (<0.1% total solution) for single-dose assays, or 0–120 nM 1, 25D3 for concentration-response curves. Luciferase activity was measured 24 hours after the exposure using the Dual-Glo luciferase assay system (Promega) following the manufacturer's protocols. Luciferase readings were normalized by cotransfection with pRL-CMV, and VDR response was normalized to an empty pSG5 vector control. To compare VDR transactivation in the presence and absence of coregulators, VDR+ coregulator response was normalized to VDR response in the absence of coregulators. Experiments were repeated at least twice and performed in replicates of four wells. Single-dose assays were analyzed using one-way ANOVAs followed by a Tukey's honestly significant difference (HSD) post hoc test. The EC50, the 95% confidence interval, and the maximal efficacy (EMAX) of the concentration-response curves were determined via a linear regression analysis using a sigmoidal dose-response calculation with a variable slope. All statistics were conducted in GraphPad Prism 4 (GraphPad Inc).
Mammalian two-hybrid assays
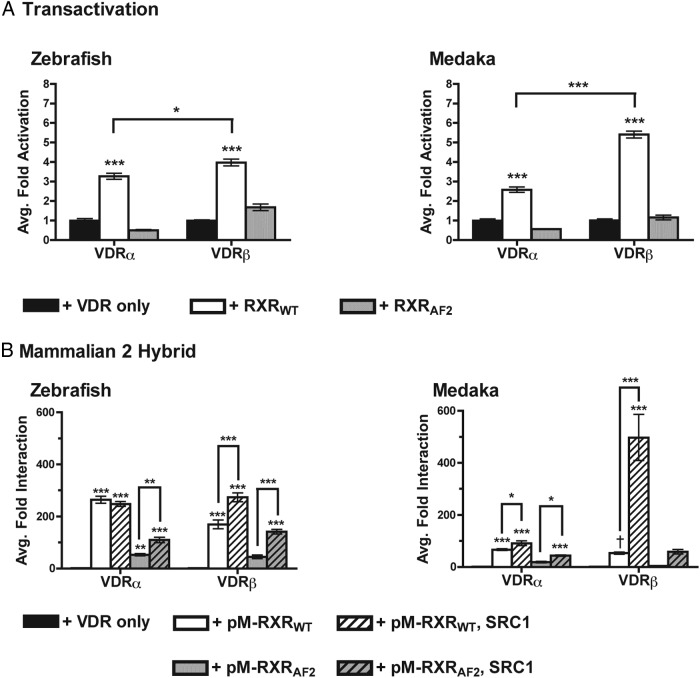
HepG2 cells were seeded into 96-well plates and transfected 24 hours later as described above. Each well contained 33.6 ng pVP16-VDR, 33.6 ng pM-coregulator as a fusion protein containing the yeast Gal4 DNA-binding domain fused to either full-length RXR (pM-RXRWT, pM-RXRAF2), or the defined NR box of one of the steroid receptor coactivator (SRC)/p160 coactivators (31): pM-SRC1241–386, pM-GRIP1479–767, or pM-ACTR392–1005. Luciferase reporters included 126.6 ng 5XGal4-TATA-Luc containing response elements for the yeast Gal4 DNA-binding domain and 3 ng pRL-CMV. Controls consisted of transfections containing empty pM, pVP16, or both empty pM and pVP16 expression vectors or ethanol as a vehicle control. Media were replaced the following day with complete MEM containing 120 nM 1, 25D3 or ethanol control. The cells were tested for luciferase activity 24 hours after the exposure as described above. VDR-coregulator interaction was normalized to VDR in the absence of a coregulator construct. Experiments were repeated at least twice and performed in replicates of four wells. Results were analyzed via one-way ANOVAs followed by Tukey's HSD post hoc test in GraphPad Prism 4.
Electrophoretic mobility shift assays
Recombinant full-length VDR and RXRWT proteins for EMSAs were expressed and purified as follows. The protein expression constructs pET32a-VDR or pET32a-RXRWT were transformed into the BL21(DE3)pLysS strain of Escherichia coli following the manufacturer's protocol (Agilent Technologies). Overnight starter cultures (10 mL) were used to inoculate 250 mL LB/amp and grown at 37°C with shaking until the OD600 was approximately 0.6. Protein expression was induced by the addition of 1 mM isopropyl-1-thio-β-D-galactopyranoside and 20 μM ZnCl2, followed by a 3-hour incubation at 25°C and 200 rpm. Cultures were centrifuged at 4000 × g for 20 minutes at 4°C, the supernatant was discarded, and the pellets were stored at −20°C overnight. The QIAexpress Ni-NTA Fast Start kit (QIAGEN) was used to lyse the bacteria and purify the recombinant His-tagged VDR and RXR proteins under native conditions following the manufacturer's protocol. Purified protein concentrations were determined using the average of three A280 measurements using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Proteins were visualized via Western blot using an anti-His antibody (number 34660; QIAGEN).
DNA-protein binding reactions were carried out using the canonical vitamin D response element (VDRE) (32) (5′-AGCTTCAGGTCAAGGAGGTCAGAGAGC-3′), a mutant form of the canonical sequence (5′-AGCTTCAGAACAAGGAGAACAGAGAGC-3′), or a VDRE containing the DR3 found within the distal promoter region of the CYP3A4-XREM reporter used in transient transfection studies (5′-GCTGAATGAACTTGCTGACCCTCTGCT-3′) (30). Single-stranded 5′-Cy5-labeled and unlabeled oligos were purchased from Integrated DNA Technologies. EMSAs were carried out in a total volume of 20 μL, containing 100 ng of recombinant VDR and 100 ng recombinant RXRWT where indicated. Receptors were incubated at 25°C for 45 minutes in a binding buffer (100 mM KCl, 10 mM HEPES, 1 mM EDTA, 0.1 mg/mL BSA, 4 μg/mL sonicated salmon sperm, 1.0 mM dithiothreitol, 1% glycerol, and 20 mM MgCl2) and 100 nM 1, 25D3 or ethanol as a vehicle control. After the first incubation, 1 pmol Cy5-labeled double-stranded VDRE was added to each reaction, and incubated for an additional 30 minutes. Competition experiments were performed in the presence of 100-fold molar excess of unlabeled wild-type or mutant VDREs. Negative controls include expressed affinity tags isolated from empty pET32a vector stocks, binding reactions run with either VDR or RXR individually, and ethanol as a vehicle control. Protein-DNA complexes were resolved on a 6% nondenaturing acrylamide gel in ice-cold 0.5× buffer (45 mM Tris base; 45 mM boric acid; 1 mM EDTA, pH 8.0) at 100 V for 90 minutes. Gels were visualized on a Storm 865 (GE Healthcare Life Sciences).
Saturation binding analysis
Recombinant VDR expression was conducted as previously described in Cos7 cells, which are void of endogenous VDR and are highly amenable to expression of heterologous nuclear receptors (25). Briefly, cells were maintained in complete DMEM supplemented with 10% fetal bovine serum and passaged every 4–5 days according to established protocols. To prepare lysates, cells were seeded at 3.0 × 106 cells per 150-mm dish, and transfected the following day with 4 μg pSG5-VDR and 4 μg pSG5-RXRWT along with 16 μg of pBSII vector as carrier DNA, using Lipofectamine 2000 (Life Technologies). Media were replaced 24 hours after the transfection, and 48 hours after the transfection, cells were washed twice with 10 mL Dulbecco's PBS and harvested by trypsinization. The cells were centrifuged for 5 minutes at 1000 rpm at 4°C and resuspended twice in 2 mL ice-cold Dulbecco's PBS. After the second resuspension, the cells were centrifuged and resuspended in 1 mL of an ice-cold buffer of 0.15 M KCl, 1 mM EDTA, 10 mM Tris HCl, 0.3 mM ZnCl2, 200× dilution protease inhibitor cocktail (EMD Millipore), and 5 mM dithiothreitol (pH 7.6). Cells were sonicated (12 1 sec bursts at 25% power) and centrifuged at 100 000 × g for 30 minutes at 4°C. The supernatant containing the lysate was divided into aliquots and stored at −80°C.
The affinity of 1, 25D3 for teleost VDRα and VDRβ was assessed as follows: lysate was diluted 1:20 in an ice-cold buffer of 0.15 M KCl, 1 mM EDTA, 10 mM Tris HCl, 0.3 mM ZnCl2, 200× dilution protease inhibitor cocktail (EMD Millipore), and 5 mM dithiothreitol (pH 7.6). The radioligand [3H]-1, 25D3 [1, 25-(OH)2-26, 27-[3H]dimethyl-vitamin D3, original specific activity 157 Ci/mmol; PerkinElmer] was diluted to 25 Ci/mmol with unlabeled 1 μM 1, 25D3 and further diluted with ethanol to obtain the desired concentrations (0–1.0 nM). The appropriate concentration of [3H]-1, 25D3 (10 μL) was added to 200 μL lysate, shaken, and incubated overnight at 4°C. Unbound ligand was removed with the addition of 80 μL of a 0.5% dextran-2.5% charcoal suspension in a buffer of 150 mM NaCl, 15 mM NaN3, 100 mM anhydrous Na2HPO4, 39 mM NaH2PO4 · H2O, 0.1% gelatin (pH 7.6) incubated on ice for 15 minutes with brief shaking every 5 minutes. Samples were centrifuged at 5000 × g for 5 minutes at 4°C, and 200 μL of the supernatant containing the receptor-bound ligand was removed for scintillation counting. Assays were repeated at least twice, with duplicate tubes for each concentration. Total binding was determined by using lysate transfected with both pSG5-VDR and pSG5-RXRWT, and nonspecific binding was determined with lysate transfected with the empty pSG5 vector in the place of pSG5-VDR. Specific binding was obtained by subtracting the nonspecific binding counts from the total binding counts. Hyperbolic one-site binding curves were fit using GraphPad Prism 4. Reported dissociation constant (Kd) values are the average of three separate curves ± SEM. Variation between the average Kd values of each VDR was tested via a one-way ANOVA followed by Tukey's HSD post hoc test.
Results
VDR transactivation in response to 1, 25D3
To analyze differential ligand potency and efficacy, concentration-response assays were conducted using a range of 1, 25D3 concentrations between 0 and 120 nM, using transactivation as an end point. Results demonstrate that EC50 values were consistently in the low nanomolar range for all VDRs tested, and no significant difference between paralogs or between species was detected (P = .6301) (Figure 1). Conversely, significant variation in EMAX levels was observed between both species and VDRα and VDRβ paralogs (P < .0001), with medaka and zebrafish VDRβ exhibiting significant attenuation in response compared with their corresponding VDRα paralogs (Figure 1).
Figure 1.
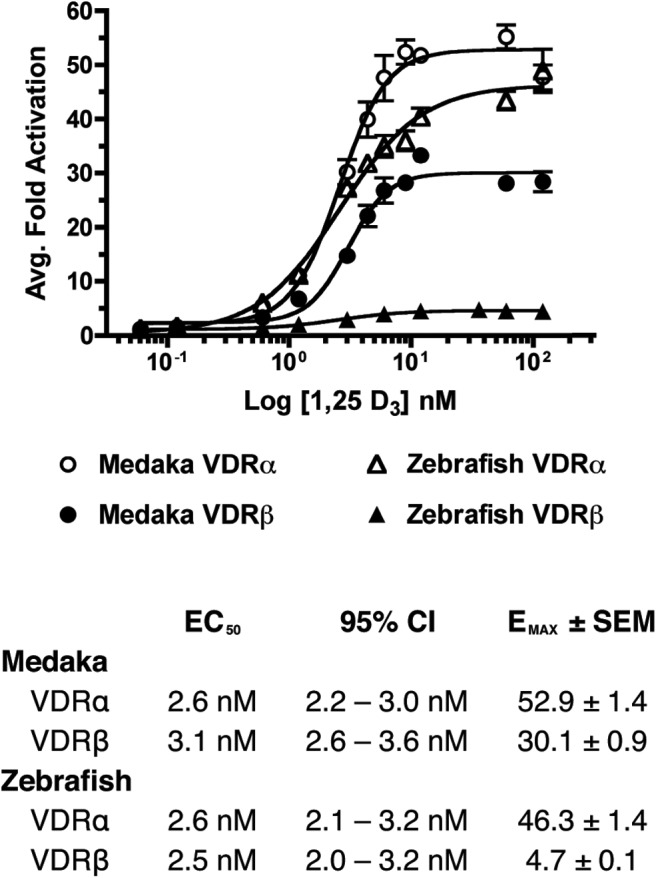
Concentration-response curves for the transactivation of medaka (circles) and zebrafish (triangles) VDRα (open) and VDRβ (closed) in response to 0–120 nM 1, 25 D3. HepG2 cells were transfected with pSG5-VDR, XREM-Luc, and pRL-CMV as an internal control as described in Materials and Methods. Data are represented as the average fold activation normalized to the vehicle control ± SEM (n = 4).
Saturation binding analysis
Innate ligand binding kinetics for each VDR paralog were determined using saturation binding analysis with radiolabeled [3H]-1, 25D3 (Figure 2). The Kd values were obtained for each VDR and were all within the subnanomolar range (10−10 M), demonstrating high-affinity binding between 1, 25D3 and each VDR paralog/ortholog tested. There was no observed statistically significant difference in ligand affinities between VDR paralogs or between species (P = .6198).
Figure 2.
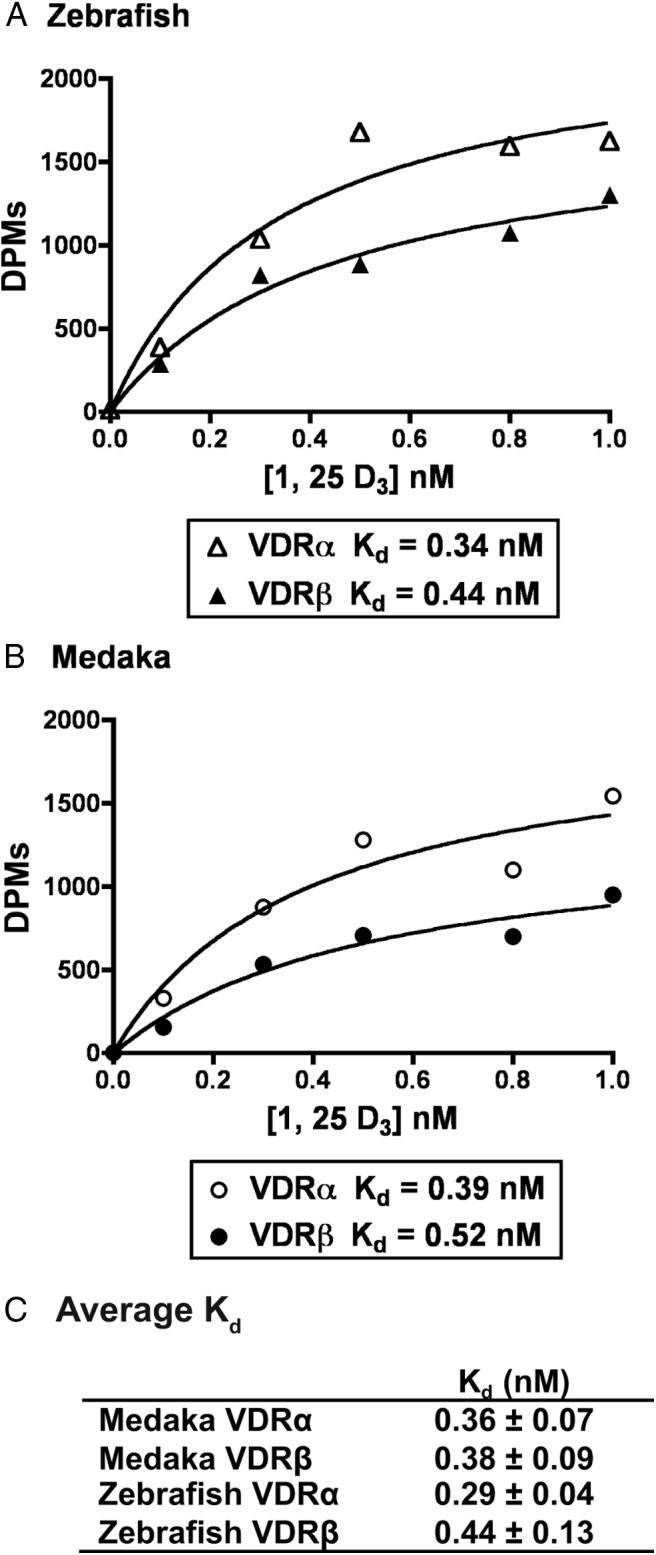
Saturation binding analysis of [3H]-1, 25 D3. Zebrafish VDRα and VDRβ (A) and medaka VDRα and VDRβ (B) are shown. Lysates were prepared from transfected Cos7 cells and incubated with increasing concentrations of [3H]-1, 25D3 as described in Materials and Methods. Unbound ligand was removed as described. C, The reported Kd for each VDR is the average of three separate experiments ± SEM. Panels A and B are specific binding data from a representative experiment.
DNA binding
EMSAs were conducted to test the protein-DNA interactions between VDR paralogs and well-defined VDREs. For all VDRs tested, a receptor-DNA binding complex was observed only in the presence of RXRWT (Figure 3A, odd lanes vs even lanes). No binding complex was observed when VDR or RXRWT was used individually, indicating a dependence on VDR-RXR heterodimerization for DNA binding and a definitive lack of VDR or RXRWT binding as putative homodimers. To ensure the specificity of the reaction, all EMSAs were repeated in the presence of excess unlabeled wild-type or mutant VDREs (Figure 3B for canonical VDRE and Figure 3C for XREM VDRE). Competition assays using wild type competitors were effectively able to outcompete the VDR-RXRWT binding to both the canonical VDRE (Figure 3B, lane 1 vs 3, and lane 4 vs 6) and XREM VDRE (Figure 3C, lanes 1 vs 2 and 3 vs 4 for VDRα and lanes 5 vs 6 and 7 vs 8 for VDRβ). Competition assays using an unlabeled mutant VDRE had no effect on VDR binding to the canonical VDRE (Figure 3B, lane 1 vs 2 and lane 4 vs 5).
Figure 3.
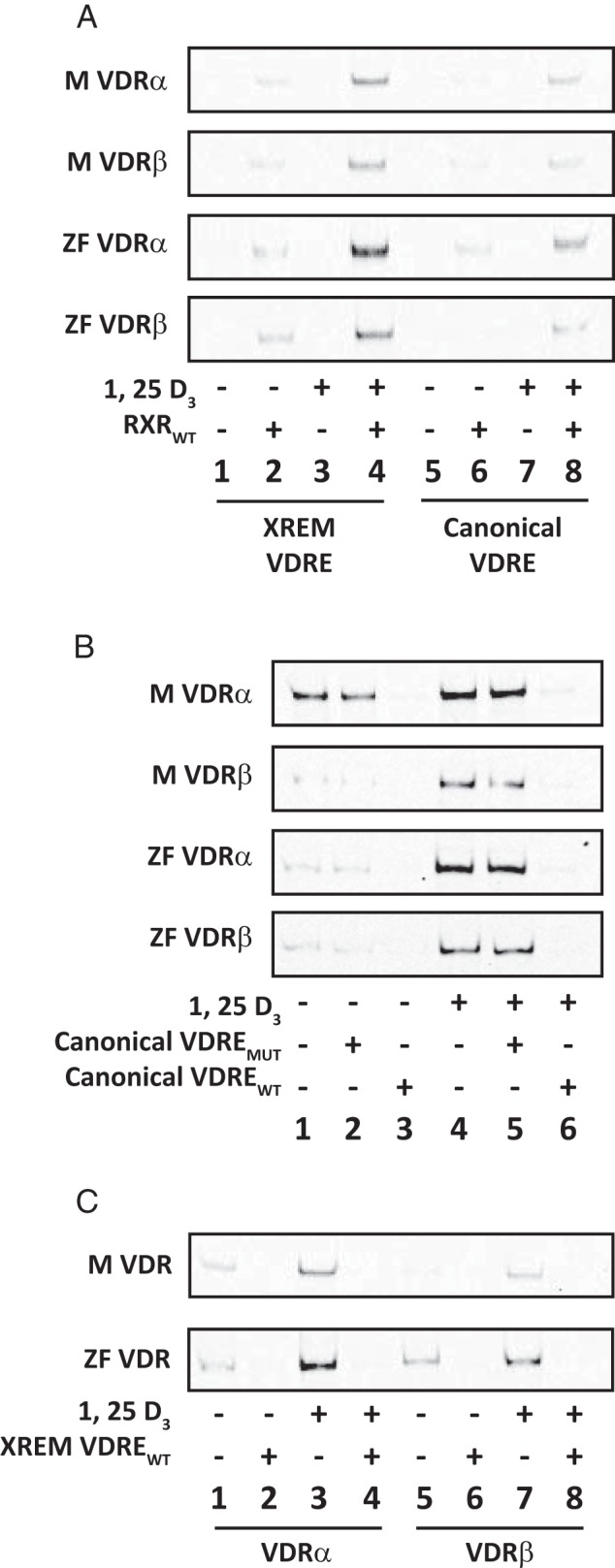
EMSAs of recombinant VDRα and VDRβ binding to canonical and noncanonical VDREs. A, Medaka (M) and zebrafish (ZF) VDRα and VDRβ were analyzed for their ability to form DNA binding complexes with both canonical (lanes 5–8) and noncanonical XREM (lanes 1–4) VDREs. Receptors were analyzed both in the presence (+) and absence (−) of recombinant RXRWT and 100 nM 1, 25 D3. B, Competition assays with the canonical VDRE using both unlabeled mutant VDRE (canonical VDREMUT) and unlabeled wild-type VDRE (canonical VDREWT) as competitors. Assays were run both in the absence (lanes 1–3) and presence (lanes 4–6) of 100 nM 1, 25D3. Lanes 1 and 4 demonstrate DNA binding in the absence of either competitor. Lanes 2 and 5 include 100-fold molar excess of the mutant competitor, and lanes 3 and 6 include 100-fold molar excess of the wild-type competitor. C, Competition assays with the divergent XREM VDRE using an unlabeled wild-type competitor VDRE (XREM VDREWT). Lanes 1–4 represent VDRα, and lanes 5–8 represent VDRβ for medaka (M) and zebrafish (ZF). Assays were run both in the presence (lanes 3 and 4 for VDRα and lanes 7 and 8 for VDRβ) and absence (lanes 1 and 2 for VDRα and lanes 5 and 6 for VDRβ) of 100 nM 1, 25 D3. Even numbered lanes include 100-fold molar excess of the wild-type competitor, whereas odd numbered lanes demonstrate DNA binding in the absence of the competitor.
Partial VDR-DNA binding was observed in the absence of 1, 25D3 (Figure 3A, lanes 2 and 6; 3B, lanes 1 and 2; 3C, lanes 1 and 5). However, protein-DNA interaction with all VDREs tested was enhanced in the presence of 1, 25D3 ligand (Figure 3A, lanes 2 vs 4 and lanes 6 vs 8; 3B, lanes 1 vs 4; 3C, lanes 1 vs 3 for VDRα and lanes 5 vs 7 for VDRβ). VDRα paralogs exhibited enriched protein-DNA interactions compared with the VDRβ paralogs with both the canonical VDRE and distal XREM VDRE (Figure 3, A–C).
Coregulator interaction: heterodimerization with RXR
A series of studies were conducted to determine whether VDR transactivation is significantly enhanced by the presence of supplemented RXR. In transient transactivation studies, the addition of full-length RXRWT resulted in a significant increase in transactivation of all VDRs compared with the activity of VDR alone (Figure 4A, P < .001 for all). Medaka and zebrafish VDRβ exhibited a greater increase in transactivation compared with their respective VDRα paralogs. In each instance in which the RXRAF2 mutant was substituted for RXRWT, luciferase activity was significantly attenuated as expected compared with studies with RXRWT.
Figure 4.
Assessment of VDR-RXR interactions. A, Analysis of overexpressed RXR on VDR transactivation in response to 120 nM 1, 25-D3. Data are represented as the average fold activation normalized to VDR alone (no RXR) ± SEM (n = 4). Asterisks above bars represent a significant increase in VDR transactivation in the presence of RXR compared to the control (no RXR). ***, P < .001; **, P < .01; *, P < .05. Asterisks above the brackets indicate a significant difference between VDRα and VDRβ. B, Mammalian two-hybrid analysis of VDR-RXR interaction in response to 120 nM 1, 25 D3. Data are represented as the average fold interaction normalized to VDR alone (no pM-RXR). Asterisks above bars represent either a significant interaction between VDR and RXR compared with the control. ***, P < .001; **, P < .01; *, P < .05. Asterisks above brackets indicate a significant differences in VDR-RXR interaction in the presence and absence of SRC1. The symbol (dagger) indicates that the interaction between medaka VDRβ and RXR was found to be significant with an unpaired t test compared with VDRβ alone: t6 = 13.71, P < .0001.
VDR-RXR heterodimerization was further confirmed through mammalian two-hybrid assays (M2H). Identical assays were conducted using the mutant RXRAF2. Data from these studies demonstrate a direct interaction between each teleost VDR tested and pM-RXRWT in the presence of 1, 25D3 (Figure 4B). Protein-protein interactions were not observed in the absence of ligand, and VDR-RXR interaction was significantly attenuated with the substitution of pM-RXRWT with pM-RXRAF2, highlighting the necessity of the C terminal AF2 domain of RXR for VDR transactivation. The addition of SRC1 significantly increased the interaction between RXRWT and VDRβ for both species. In contrast, a much smaller increase in interaction was observed between RXRWT and medaka VDRα in the presence of SRC1, whereas SRC1 did not have an effect on zebrafish VDRα and RXRWT.
Coregulator interaction: SRC/p160 family of NR coactivators
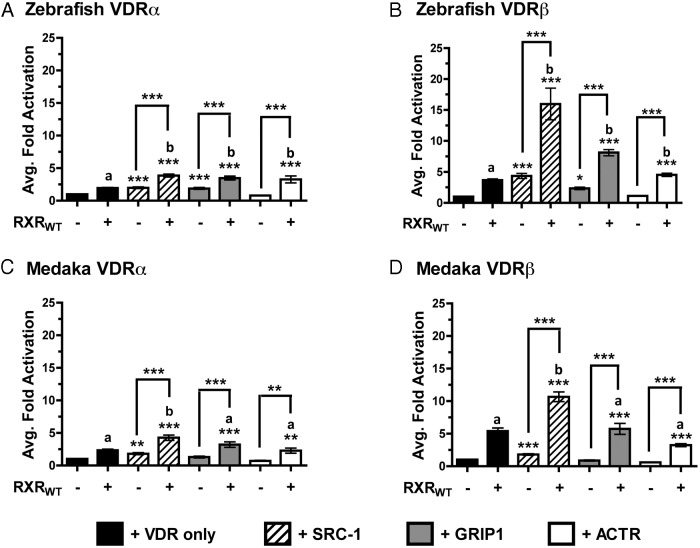
Protein interaction studies were expanded to include the SRC/p160 family of NR coactivators. This family includes SRC1, glucocorticoid receptor interacting protein-1 (GRIP1), and activator of thyroid and retinoic acid receptor (ACTR). Cotransfection of the SRC/p160 coactivators in transient transactivation assays had varying effects on the VDR paralogs. SRC1 significantly increased VDR activation for each VDR tested (Figure 5, A–D). GRIP1 enhanced only zebrafish VDRα and VDRβ activation but did not affect medaka VDRα or VDRβ. ACTR proved ineffective, exhibiting no enhancement of VDR transactivation.
Figure 5.
VDR transactivation with supplemented SRC/p160 coactivators: analysis of VDR transactivation in the presence of overexpressed SRC1, GRIP1, and ACTR, both in the presence (+) and absence (−) of overexpressed RXRWT in response to 120 nM 1, 25D3. Data are represented as the average fold activation normalized to VDR alone ± SEM (n = 4). Asterisks above the bars indicate a significant increase in VDR transactivation compared with the VDR alone control. ***, P < .001; **, P < .01; *, P < .05. Asterisks above the brackets indicate a significant difference in transactivation between the presence and absence of RXRWT. The letter b indicates cotransfection with both RXRWT and SRC/p160 coactivators had a greater effect on VDR transactivation compared with VDR + RXRWT in the absence of an SRC/p160 construct (black bar). The letter a indicates that no difference in VDR transactivation was observed in the presence of both RXRWT and the indicated SRC coactivator vs. VDR + RXRWT in the absence of the SRC/p160 coactivator.
Cotransfection of both RXRWT and the SRC/p160 coactivators significantly enhanced the activation of each VDR tested in the transactivation studies (Figure 5). The level of transactivation achieved for all four VDRs tested were greater with the combination of both RXRWT and SRC1 compared with either coactivator used individually. However this combination of coregulators appeared to have a preferential effect on VDRβ activation compared with VDRα for both medaka and zebrafish. The enhanced VDR transactivation was additionally demonstrated with the cotransfection of RXRWT combined with GRIP1 or ACTR in zebrafish; however, these combinations of coregulators did not augment the transactivation of medaka VDR paralogs (Figure 5).
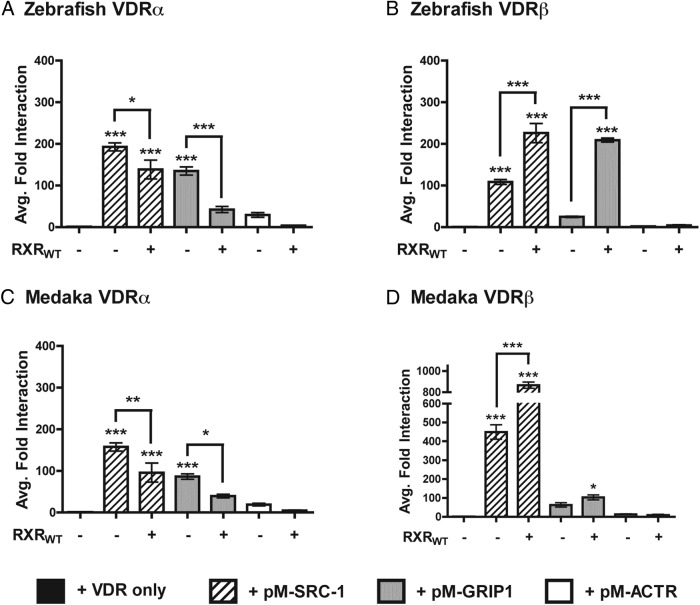
Mammalian two-hybrid assays were conducted to assess the protein-protein interaction between VDR and individual SRC/p160 coactivators (Figure 6, A–D). Given that the cotransfection of both RXRWT and SRC/p160 coactivators significantly enhanced VDR transactivation, M2H assays were conducted in the presence and absence of RXRWT to determine whether the obligate heterodimer partner facilitates the interaction between VDR and the SRC/p160 coactivators. First, M2H assays conducted in the absence of RXRWT demonstrate that both VDRα and VDRβ from both species maintain a strong and significant interaction with SRC1, suggesting that both VDR paralogs directly recruit SRC1 in response to 1, 25D3 (Figure 6, A–D). Comparatively, only VDRα from both species demonstrated a significant interaction with GRIP1 in the absence of RXRWT (Figure 6, A and C vs B and D). No protein-protein interactions were observed between ACTR and any of the VDRs tested. The addition of RXRWT to the SRC/p160 M2H assays had contrasting effects on coactivator recruitment. RXRWT significantly enhanced VDRβ interaction with both SRC1 and GRIP1, consistent with that observed in transient transactivation assays. Conversely, addition of RXRWT to the SRC/p160 M2H assays with medaka and zebrafish VDRα resulted in an attenuated response compared with the same assay in the absence of RXRWT (Figure 6, A vs B and Figure 6, C vs D).
Figure 6.
Analysis of VDR recruitment of members of the SRC/p160 family of NR coactivators in response to 120 nM 1, 25D3. Data are represented as the average fold interaction ± SEM (n = 4). Data are normalized to VDR in the absence of coregulators. Asterisks above the bars indicate a significant interaction between VDR and SRC-1, GRIP1, or ACTR compared with the VDR alone control. ***, P < .001; **, P < .01; *, P < .05. Asterisks above the brackets indicate a significant difference in VDR-SRC/p160 interaction in the presence and absence of RXRWT.
Discussion
The duplication and divergence of transcriptional regulators is thought to have played an essential role in the evolution of vertebrate complexity and diversity. Mining of teleost genomes has identified numerous instances of duplicate and paralogous transcriptional regulators. After the discovery of VDRβ, it was determined that it is paralogous to VDRα and represents a novel subfamily of ligand-activated transcription factors within the vertebrate NR1I family (15, 22, 23). To date, however, molecular function and physiological significance of duplicate teleost NRs is based predominantly on structural similarities and orthology to their mammalian counterparts.
Using transient transactivation assays, we first confirmed that VDRα and VDRβ from medaka and zebrafish are capable of transactivation in response to the endogenous VDR ligand, 1, 25D3. By titrating the concentration of 1, 25D3, we established an EC50 value for each VDR. EC50 values were highly similar between each VDR tested, indicating that 1, 25D3 is a highly potent ligand both between (orthologs) and within (paralogs) species. These data are consistent with that observed from other teleost VDR studies that used Gal4-VDRLBD chimeras to assess VDR transactivation across phylogenetically diverse taxa (22, 33). All VDRs examined to date exhibit at least modest transactivation and comparable EC50 values with 1, 25D3, including the sea lamprey, a primitive vertebrate (25). This suggests that the association between VDR and 1, 25D3 occurred early in vertebrate evolution and was highly conserved.
Conversely, the examination of maximal transactivational efficacy (EMAX) between the VDR paralogs of both species revealed significant differences. We sought to further establish the functional basis for these transactivational differences through examining core NR functions between VDRα and VDRβ in the two teleost species. Because ligand binding is the initiating event of NR activation, we initially investigated the possibility that the observed transactivational differences between VDRα and VDRβ were associated with differential affinity for 1, 25D3. VDRα and VDRβ from both species exhibited high affinity binding with 1, 25D3, similar to previously reported values for mammalian VDR proteins. However, statistically significant differences in ligand affinities were not observed between paralogs or species, indicating that differential ligand affinities are likely not the root cause of the observed transactivational disparities. In fact, it is well known that ligand affinity and transactivation efficacy of specific NR ligands are not proportional (34). This phenomenon is likely due to the ability of ligands to induce subtle variations in receptor conformation that elicit different biological responses such as DNA binding and protein-protein interactions. High ligand affinity but low transactivational efficacy of VDR has been observed previously in other aquatic vertebrates. For example, the sea lamprey (Petromyzon marinus) VDR exhibits high affinity binding with 1, 25D3 (Kd = 10−10 M); however, transactivation of this VDR with 1, 25D3 is minimal (<4.0-fold) (25).
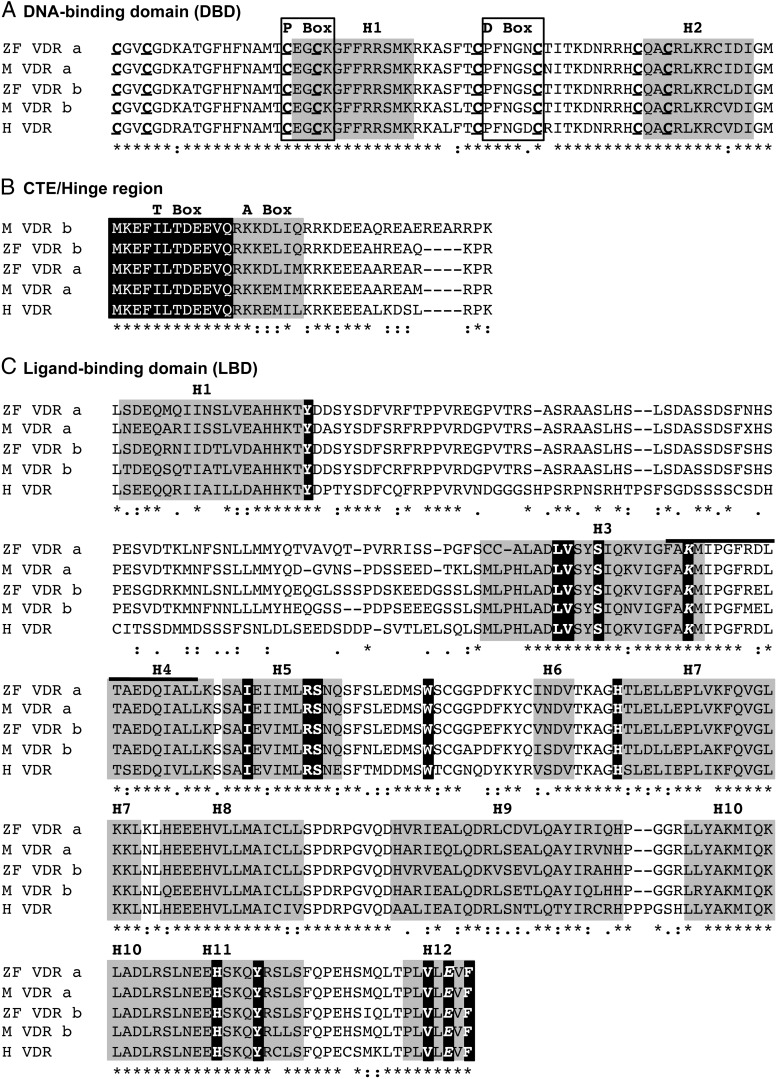
We next examined whether differences in VDRα and VDRβ transactivation were due to modified NR functions that occur subsequent to ligand binding. We began by assessing the ability of the VDRs to bind to well-characterized VDREs. Canonical VDREs are comprised of a direct-repeat of two hexameric half sites separated by a spacer of three nucleotides (referred to as a DR3). Our results demonstrate that distinct differences in DNA binding occur between the VDRα and VDRβ paralogs, with VDRα exhibiting preferential binding compared with VDRβ. DNA binding differences were conserved between species and occurred with both canonical and noncanonical VDREs. Sequence similarities suggest that the observed DNA binding differences between VDRα and VDRβ are likely not related to sequence divergence in the DNA binding domain (DBD). The DBD sequences of the four VDR paralogs are highly homologous, sharing 95%–96% amino acid sequence similarity (Figure 7A). All eight cysteine residues essential for the zinc finger structure and the recognition helix are identical between VDRα and VDRβ and human VDR (35–37).
Figure 7.
Comparison of VDR paralog functional domains. Amino acid sequence alignment of medaka (M) and zebrafish (ZF) VDRα and VDRβ. Human VDR (H) is included for comparison. Predicted amino acid sequences were aligned using ClustalW. Functional domains and domain elements were determined based on previous work with human VDR discussed in the manuscript. Fully conserved residues are indicated by an asterisk. Partial conservation is indicated by a colon (strong) or period (weak). Lack of residue consensus is indicated by a blank space. For the DNA-binding domain (A), residues that make up the P-box and D-box are indicated with a black box, and helix 1 (the recognition helix) and helix 2 are highlighted in grey. Conserved cysteine residues involved in zinc finger structure are both bold and underlined. For the CTE/hinge region (B), the T-box is highlighted in black with white letters, whereas the A-box is highlighted in grey with black letters. For the ligand-binding domain (C), the estimated α-helices are highlighted in grey. Amino acid residues that serve as contacts for 1, 25 D3 are indicated in white letters highlighted in black. The residues involved in the charge clamp formation are both bold and italicized. The E1 region within H3 and H4 is indicated with a solid black line above the alignment that includes part of H3 and H4.
Additionally, studies have demonstrated the importance of residues in the C-terminal extension (CTE) for sufficient DNA binding and transactivation (38) (Figure 7B). These residues likely increase the affinity of VDR-DNA interaction by aiding in nonspecific interaction within the minor grove of the DNA backbone (T-box) and impart additional RXR dimerization and sequence specificity to the conserved DBD core region (A-box) (39, 40). Although more sequence variation is observed in the CTE region between VDRα and VDRβ, the basic residues implicated in DNA binding are conserved across all medaka and zebrafish VDR paralogs (32). Mutation studies have also demonstrated that modifications within the LBD may impact DNA binding through alterations in RXR heterodimerization (41, 42). Our DNA binding studies demonstrate a stringent requirement for RXR for binding complex formation. In addition, we demonstrate that DNA binding is significantly enhanced with addition of 1, 25D3. This suggests that the presence of ligand may favorably alter the conformation of the VDR LBD, resulting in enhanced VDR-RXR interactions and facilitating the interaction with the VDRE (43–45).
To test the impact of RXR on VDR transactivational efficacy, either wild-type RXR (RXRWT) or a truncated RXR mutant (RXRAF2) lacking the C-terminal ligand-dependent AF2 region was cotransfected with VDR in transient transactivation studies. We consistently observed an increase in VDR transactivational efficacy in the presence of RXRWT. The use of the RXRAF2 mutant attenuated VDR transactivation to background levels and significantly attenuated the interaction between VDR and RXR in M2H studies. Previous studies have demonstrated that the AF2 region of both VDR and RXR are required for vitamin D-dependent gene transcription (46, 47). We speculate that the partial loss of a response with RXRAF2 is likely due to either an inability to recruit necessary coactivators due to the loss of the charge clamp or to corepressor interactions on sites previously shielded by the RXR AF2 (48). In conjunction with DNA binding data, these results support obligate interactions between both NRs for essential VDR functions in both medaka and zebrafish paralogs.
In addition to RXR, we also studied the effects of the SRC/p160 family of nuclear receptor coactivators on VDR transactivation. Members of the SRC/p160 family directly interact with the AF2 region of NRs and enhance NR-mediated transcription of target genes by both chromatin modification and the recruitment of additional proteins to the transcription complex (31). Although the partnership between VDR and RXR appears to be well conserved, dramatic functional differences between VDRα and VDRβ were observed with the cotransfection of both RXRWT and the SRC/p160 coactivators in transient transactivation assays. Specifically we observed preferential transactivation of VDRβ paralogs over VDRα in the presence of overexpressed SRC1 and RXRWT in both species. This transactivational increase was significantly greater than the individual effects of either coregulator used singularly. In addition, results from our M2H assays further support the notion of preferential effects of RXRWT and SRC1 on VDRβ transactivation.
Examination of key regions within the VDR known for protein-protein interaction with RXR and the SRC/p160 coactivators, including helices 9 and 10, the AF2 and E1 region, and the T box of the CTE, all demonstrate a high degree of conservation between VDRα and VDRβ paralogs (Figure 7). Furthermore, amino acid variation within these regions is usually not conserved in both VDRα and VDRβ paralogs, suggesting that the observed variation may not explain the paralog-specific functional differences. Second, the fact that both VDRα and VDRβ bind 1, 25D3 with equal affinity indicates that differential ligand affinities are not driving differences in protein interactions between paralogs. Rather, we speculate that receptor-ligand interactions between VDR and 1, 25D3 may result in different molecular conformations in VDRα vs VDRβ that can differentially influence downstream functions. Mammalian VDR bound to 1, 25D3 produces a significant conformational change in the receptor from the inactive apo to the active holo state (49). This conformational change appears to be consistent across all mammalian species tested thus far (50, 51) and crystal studies with zebrafish VDRα bound to both 1, 25D3 and Gemini (a synthetic VDR agonist), reveal a highly similar active conformation compared with human VDR (52). This supports our data indicating a high degree of functional similarity between the two species and provides an initial indication that the VDRα paralog more closely resembles the terrestrial ortholog of this nuclear receptor. Crystal structures of both VDRα/β paralogs with 1, 25D3 may provide invaluable structural comparisons because the differences observed between the two paralogs may be related to conformational differences and provide additional detail on the evolution of aquatic and terrestrial forms of this important nuclear receptor.
It has additionally been demonstrated that the active conformation of VDR is ligand specific. Jurutka et al (53) have suggested that lithocholic acid induces an alternative active conformation when bound to VDR compared with 1, 25D3, leading to differential interactions with RXR and other coactivator proteins. These studies further demonstrate that ligand-induced conformational differences are associated with altered protein-protein interactions with coactivators and heterodimerization partners due to suboptimal positioning of helices 9, 10, and 12 (53, 54). These diminished protein-protein interactions ultimately result in significantly reduced VDR transactivation. Although 1, 25D3 was the only ligand in our study, we speculate that subtle conformational differences between VDR paralogs may lead to the altered coregulator interactions described above. Specifically, the active conformation induced by 1, 25D3 in VDRβ may be less stable compared with VDRα, resulting in attenuated affinity for RXR and the SRC/p160 coactivators under nonsaturating coactivator conditions. However, enhanced activity in transient transactivation studies and enhanced protein interactions observed with VDRβ may be due to a compensatory effect of overexpressed coregulators. Analogous to our observations, compensatory effects of overexpressed coregulators have been previously observed with a VDRR391C mutant associated with a form of hereditary vitamin D-resistant rickets (43, 47).
Two possible models may explain the compensatory mechanism of overexpressed RXR and SRC1 on VDR transactivation. One mechanism involves the AF2 regions of both VDR and RXR interacting with different LXXLL motifs of the same SRC/p160 coactivator (47). This bridging effect may help stabilize a less optimal heterodimer. The pattern of one coactivator per heterodimer has been previously observed with other NR heterodimers such as RXR-RAR (55). Alternatively, a second model suggests that each heterodimer partner interacts with a distinct and separate coactivator. A study by Yang et al (56) found that coactivator recruitment to the permissive peroxisomal proliferator-activated receptor-γ-RXR heterodimer is highly ligand specific. For instance, the presence of LG268, an RXR-specific ligand, resulted in protein-protein interaction only between SRC-1 and RXR and not peroxisomal proliferator-activated receptor-γ. Differential coactivator recruitment between heterodimer partners may potentially explain our M2H data with VDRα. The fact that the addition of RXRWT appears to attenuate VDRα-SRC1 interactions in M2H assays suggests that RXRWT may be sequestering the SRC1 proteins, such that they are effectively no longer stoichiometrically proportional for the assay.
In summary, we have identified evidence of a functional divergence between VDRα and VDRβ paralogs in teleost fish. Our results suggest that differences in protein-protein interactions between the VDR paralogs and essential coregulators may be driving the observed differential ligand sensitivities between VDRα and VDRβ. Furthermore, this divergence occurred early in teleost evolution because the observed functional differences are highly conserved in two distantly related species. Further studies using VDRs cloned from species that diverged before the 3R event are necessary to gain a better understanding of derived and ancestral VDR functions of the VDR paralogs. Additional studies may also aid in identifying specific evolutionary processes that lead to the observed functional differences.
Acknowledgments
This work was supported by National Science Foundation Grant IOS0818799 (to S.W.K.); a Stan and Judy Fellowship from the Mount Desert Island Biological Laboratory (to E.M.K.); a Dissertation Completion Grant from North Carolina State University (to E.M.K.); and National Institute of Environmental Health Sciences Training Grant T32-ES007046 (to E.M.K.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ACTR
- activator of thyroid and retinoic acid receptor
- AF2
- activation function-2
- 1
- 25D3, 1, 25-dihydroxyvitamin D3
- DBD
- DNA binding domain
- EMAX
- maximal efficacy
- GRIP1
- glucocorticoid receptor interacting protein-1
- HSD
- honestly significant difference
- Kd
- dissociation constant
- NR
- nuclear receptor
- RXR
- retinoid X receptor
- SRC
- steroid receptor coactivator
- VDR
- vitamin D receptor
- VDRE
- vitamin D response element
- WGD
- whole-genome duplication.
References
- 1. Dehal P, Boore J. Two rounds of whole genome duplication in the ancestral vertebrate. Plos Biol. 2005;3(10):1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crow K, Wagner G. What is the role of genome duplication in the evolution of complexity and diversity? Mol Biol Evol. 2006;23(5):887–892. [DOI] [PubMed] [Google Scholar]
- 3. Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16(7):805–814. [DOI] [PubMed] [Google Scholar]
- 4. Van de Peer Y, Maere S, Meyer A. Opinion: the evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10(10):725–732. [DOI] [PubMed] [Google Scholar]
- 5. Taylor J, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. [DOI] [PubMed] [Google Scholar]
- 6. Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17(5):699–717. [DOI] [PubMed] [Google Scholar]
- 7. Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7(5):R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoegg S, Brinkmann H, Taylor J, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59(2):190–203. [DOI] [PubMed] [Google Scholar]
- 9. Crow K, Stadler P, Lynch V, Amemiya C, Wagner G. The “fish-specific” Hox cluster duplication is coincident with the origin of teleosts. Mol Biol Evol. 2006;23(1):121–136. [DOI] [PubMed] [Google Scholar]
- 10. Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005;27(9):937–945. [DOI] [PubMed] [Google Scholar]
- 11. Nelson JS. Fishes of the World. 4th ed New York: J. Wiley; 2006. [Google Scholar]
- 12. Nadeau J, Sankoff D. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics. 1997;147(3):1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Postlethwait J, Amores A, Cresko W, Singer A, Yan Y. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20(10):481–490. [DOI] [PubMed] [Google Scholar]
- 14. Laudet V, Gronemeyer H. The Nuclear Receptor Factsbook. San Diego: Academic Press; 2002. [Google Scholar]
- 15. Maglich J, Caravella J, Lambert M, Willson T, Moore J, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31(14):4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Metpally RPR, Vigneshwar R, Sowdhamini R. Genome inventory and analysis of nuclear hormone receptors in Tetraodon nigroviridis. J Biosci. 2007;32(1):43–50. [DOI] [PubMed] [Google Scholar]
- 17. Bertrand S, Thisse B, Tavares R, et al. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genetics. 2007;3(11):e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17(10):554–556. [DOI] [PubMed] [Google Scholar]
- 19. Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21(10):1923–1937. [DOI] [PubMed] [Google Scholar]
- 20. Howarth DL. Characterization of FXR α in medaka and its involvement in hepatobiliary injury. Dissertation, Integrated Toxicology and Environmental Health Program, Duke University; 2009. [Google Scholar]
- 21. Li YC. Cloning and characterization of the vitamin D receptor from Xenopus laevis. Endocrinology. 1997;138(6):2347–2353. [DOI] [PubMed] [Google Scholar]
- 22. Howarth DL, Law SHW, Barnes B, et al. Paralogous vitamin D receptors in teleosts: transition of nuclear receptor function. Endocrinology. 2008;149(5):2411–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki T, Suzuki N, Srivastava A, Kurokawa T. Identification of cDNAs encoding two subtypes of vitamin D receptor in flounder, Paralichthys olivaceus. Biochem Bioph Res Commun. 2000;270(1):40–45. [DOI] [PubMed] [Google Scholar]
- 24. Krasowski MD, Ai N, Hagey LR, et al. The evolution of farnesoid X, vitamin D, and pregnane X receptors: insights from the green-spotted pufferfish (Tetraodon nigriviridis) and other non-mammalian species. BMC Biochem. 2011;12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitfield G, Dang H, Schluter S, et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology. 2003;144(6):2704–2716. [DOI] [PubMed] [Google Scholar]
- 26. Volff JN. Genome evolution and biodiversity in teleost fish. Heredity (Edinb). 2005;94(3):280–294. [DOI] [PubMed] [Google Scholar]
- 27. Lin CH, Su CH, Tseng DY, Ding FC, Hwang PP. Action of vitamin D and the receptor, VDRa, in calcium handling in zebrafish (Danio rerio). Plos One. 2012;7(9):e45650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rozen S, Skaletsky H. Primer3 on the WWW for General Users and for Biologist Programmers. In: Misener S, Krawetz SA, eds. Methods in Molecular Biology, vol 132: Bioinformatics Methods and Protocols. Totowa, NJ: Humana Press Inc; 1999:365–386. [DOI] [PubMed] [Google Scholar]
- 30. Drocourt L, Pascussi J, Assenat E, Fabre J, Maurel P, Vilarem M. Calcium channel modulators of the dihydropyridine family are human pregnane X receptor activators and inducers of CYP3A, CYP2B, and CYP2C in human hepatocytes. Drug Metab Dispos. 2001;29(10):1325–1331. [PubMed] [Google Scholar]
- 31. Leo C, Chen J. The SRC family of nuclear receptor coactivators. Gene. 2000;245(1):1–11. [DOI] [PubMed] [Google Scholar]
- 32. Shaffer P, Gewirth D. Vitamin D receptor-DNA interactions. Vitam Horm. 2004;68:257–273. [DOI] [PubMed] [Google Scholar]
- 33. Reschly EJ, Bainy ACD, Mattos JJ, et al. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol Biol. 2007;7:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peleg S, Liu YY, Reddy S, Horst RL, White MC, Posner GH. A 20-epi side chain restores growth-regulatory and transcriptional activities of an A ring-modified hybrid analog of 1α, 25-dihydroxyvitamin D3 without increasing its affinity to the vitamin D receptor. J Cell Biochem. 1996;63(2):149–161. [DOI] [PubMed] [Google Scholar]
- 35. Freedman LP, Towers TL. DNA binding properties of the vitamin D3 receptor zinc finger region. Mol Endocrinol. 1991;5(12):1815–1826. [DOI] [PubMed] [Google Scholar]
- 36. Liao J, Ozono K, Sone T, McDonnell DP, Pike JW. Vitamin D receptor interaction with specific DNA requires a nuclear protein and 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci USA. 1990;87(24):9751–9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Towers TL, Luisi BF, Asianov A, Freedman LP. DNA target selectivity by the vitamin D3 receptor: mechanism of dimer binding to an asymmetric repeat element. Proc Natl Acad Sci USA. 1993;90(13):6310–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsieh JC, Whitfield GK, Oza AK, et al. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry. 1999;38(49):16347–16358. [DOI] [PubMed] [Google Scholar]
- 39. Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26(6):384–390. [DOI] [PubMed] [Google Scholar]
- 40. Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375(6528):203–211. [DOI] [PubMed] [Google Scholar]
- 41. Jin CH, Kerner SA, Hong MH, Pike JW. Transcriptional activation and dimerization functions in the human vitamin D receptor. Mol Endocrinol. 1996;10(8):945–957. [DOI] [PubMed] [Google Scholar]
- 42. Nakajima S, Hsieh JC, MacDonald PN, et al. The C-terminal region of the vitamin D receptor is essential to form a complex with a receptor auxiliary factor required for high affinity binding to the vitamin D-responsive element. Mol Endocrinol. 1994;8(2):159–172. [DOI] [PubMed] [Google Scholar]
- 43. Whitfield GK, Selznick SH, Haussler CA, et al. Vitamin D receptors from patients with resistance to 1,25-dihydroxyvitamin D3: point mutations confer reduced transactivation in response to ligand and impaired interaction with the retinoid X receptor heterodimeric partner. Mol Endocrinol. 1996;10(12):1617–1631. [DOI] [PubMed] [Google Scholar]
- 44. MacDonald PN, Dowd DR, Nakajima S, et al. Retinoid X receptors stimulate and 9-cis retinoic acid inhibits 1,25-dihydroxyvitamin D3-activated expression of the rat osteocalcin gene. Mol Cell Biol. 1993;13(9):5907–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson PD, Jurutka PW, Haussler CA, Whitfield GK, Haussler MR. Heterodimeric DNA binding by the vitamin D receptor and retinoid X receptors is enhanced by 1,25-dihydroxyvitamin D3 and inhibited by 9-cis-retinoic acid. Evidence for allosteric receptor interactions. J Biol Chem. 1998;273(14):8483–8491. [DOI] [PubMed] [Google Scholar]
- 46. Bettoun D, Burris T, Houck K, et al. Retinoid X receptor is a nonsilent major contributor to vitamin D receptor-mediated transcriptional activation. Mol Endocrinol. 2003;17(11):2320–2328. [DOI] [PubMed] [Google Scholar]
- 47. Thompson PD, Remus LS, Hsieh JC, et al. Distinct retinoid X receptor activation function-2 residues mediate transactivation in homodimeric and vitamin D receptor heterodimeric contexts. J Mol Endocrinol. 2001;27(2):211–227. [DOI] [PubMed] [Google Scholar]
- 48. Schulman IG, Juguilon H, Evans RM. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16(7):3807–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rochel N, Wurtz J, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5(1):173–179. [DOI] [PubMed] [Google Scholar]
- 50. Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43(14):4101–4110. [DOI] [PubMed] [Google Scholar]
- 51. Rochel N, Moras D. Ligand binding domain of vitamin D receptors. Curr Top Med Chem. 2006;6(12):1229–1241. [DOI] [PubMed] [Google Scholar]
- 52. Ciesielski F, Rochel N, Mitschler A, Kouzmenko A, Moras D. Structural investigation of the ligand binding domain of the zebrafish VDR in complexes with 1α,25(OH)(2)D-3 and Gemini: purification, crystallization and preliminary X-ray diffraction analysis. J Steroid Biochem. 2004;89–90(1–5):55–59. [DOI] [PubMed] [Google Scholar]
- 53. Jurutka PW, Thompson PD, Whitfield GK, et al. Molecular and functional comparison of 1,25-dihydroxyvitamin D(3) and the novel vitamin D receptor ligand, lithocholic acid, in activating transcription of cytochrome P450 3A4. J Cell Biochem. 2005;94(5):917–943. [DOI] [PubMed] [Google Scholar]
- 54. Masuno H, Ikura T, Morizono D, et al. Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives. J Lipid Res. 2013;54(8):2206–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Westin S, Kurokawa R, Nolte RT, et al. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395(6698):199–202. [DOI] [PubMed] [Google Scholar]
- 56. Yang W, Rachez C, Freedman LP. Discrete roles for peroxisome proliferator-activated receptor γ and retinoid X receptor in recruiting nuclear receptor coactivators. Mol Cell Biol. 2000;20(21):8008–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]