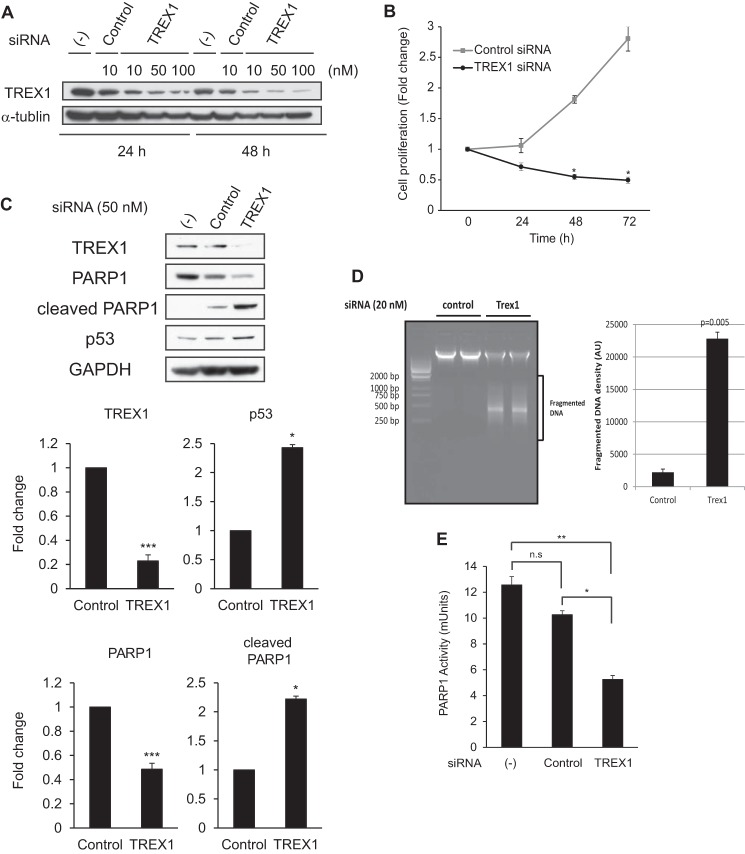
FIGURE 5.
Knockdown of TREX1 promotes PARP1 cleavage and loss of PARP1 enzymatic activity. A, HeLa cells were transfected with TREX1-specific or control siRNAs at the indicated concentrations (10–100 nm) and time course (24 or 48 h). Expression levels of endogenous TREX1 proteins were determined by Western blotting. α-Tubulin served as an endogenous loading control. (−), no siRNA treatment. B, cell proliferation assessed by the tetrazolium method was used with TREX1-specific or control siRNA-treated cells (50 nm). The cells were incubated for the indicated times (24–72 h), and the absorbance was measured for each culture using an ELISA plate reader (n = 3). Data are given as the mean ± S.E. (error bars). *, p < 0.01 by paired t test. C, HeLa cells were transfected with TREX1-specific or control siRNA (50 nm) for 48 h, and the expression levels of the indicated proteins were determined by Western blotting (n = 4). Densitometric analyses of the blots are shown in the lower panel, and values are given as the mean ± S.E. (error bars) for TREX1, p53, PARP1, and cleaved PARP1 levels relative to GAPDH levels from the four independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.0005 by paired t test. (−), no siRNA treatment. D, agarose gel electrophoresis of genomic DNA isolated from cells transfected with TREX1 and control siRNA for 48 h. The intensity of fragmented DNA in the lower panel was calculated, and mean with S.D. (error bars) was plotted. The p value was calculated by comparing with the control sample using paired t test. AU, arbitrary units. E, HeLa cells were transfected with TREX1-specific or control siRNA (50 nm) for 48 h, and PARP1 activity in cell extracts (100 ng of protein) was measured by quantification of PAR from histones attached to strip wells in a 96-well format using the HT colorimetric PARP/apoptosis assay kit (n = 3). Data are given as the mean ± S.E. (error bars). *, p < 0.05; **, p < 0.005 by paired t test. (−), no siRNA treatment. n.s., not significant.