FIGURE 5.
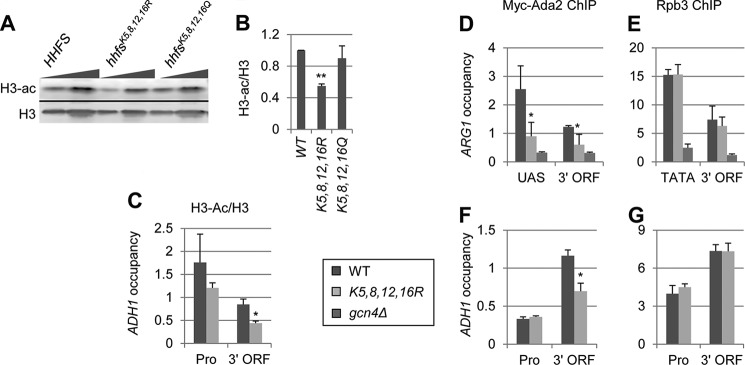
H4 acetylation stimulates H3 acetylation by SAGA. A and B, the effects of H4 tail Lys substitutions on H4 and H3 acetylation. WT and H4 mutant strains harboring the hhfsK5,8,12,16R or hhfsK5,8,12,16Q alleles (pJD62_H4_WT, Boeke-EMH-H4–172, and Boeke-EMH-H4–171, respectively) were grown to stationary phase in YPD and WCEs were subjected to Western analysis with antibodies against diacetylated H3 and the H3 C terminus. B, acetylation per histone was calculated as the ratio of H3-ac signal to the total H3 signal (n = 4). C–G, ChIP analysis of H3 and H3-ac (C), Myc-Ada2 (D and F), and Rpb3 (E and G) at ARG1 (D and E), and ADH1 (C, F, and G). WT, hhfsK5,8,12,16R, and gcn4Δ Myc-ADA2 strains (DGY655, DGY658, and DGY654) were grown to early log phase in SC at 30 °C, treated with 0.6 μm SM for 30 min, and ChIP was performed using anti-H3-ac and anti-H3 (C), anti-Myc (D and F), and anti-Rpb3 (E and G) antibodies and primers for the promoter and coding sequence of ADH1 (C, F, and G) or the enhancer and coding sequence of ARG1 (D and E). H3-ac occupancy was normalized to H3 occupancy. H3-ac/H3 and Myc-Ada2 occupancies in hhfsK5,8,12,16R cells were determined to be significantly less than in WT by Student's t test (*, p < 0.01).
