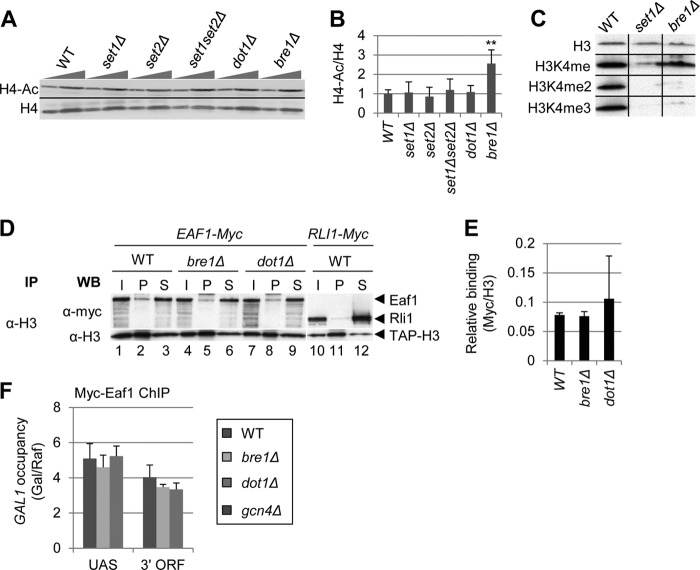
FIGURE 6.
H3K4 and K36 methylation stimulates both H4 acetylation and deacetylation. A–C, among HMT mutants, only bre1Δ alters bulk H4 acetylation. Strains BY4741, DGY183, 1257, DGY184, 4276, and 3771 were grown to stationary phase in YPD. In A, WCEs were analyzed by Western blot with anti-tetra-acetylated H4 and anti-H4 C terminus antibodies. In B, acetylation per histone was calculated as the ratio of H4-ac signal to H4 signal for each strain (n = 4). H4 acetylation in bre1Δ cells was determined to be significantly greater than WT by Student's t test (**, p < 0.001). C, Bre1 stimulates di- and trimethylation of H3K4. Strains DGY3, DGY191, and DGY427 were grown to stationary phase in YPD and WCEs were analyzed by Western blot with anti-H3K4me3, anti-H3K4me2, anti-H3K4me, and anti-H3 antibodies. D and E, coimmunoprecipitation of Myc-Eaf1 with H3 is unaltered by bre1Δ and dot1Δ. In D, strains DGY498, DGY506, DGY509, and DGY512 were grown to mid-log phase in YPD medium and WCEs were immunoprecipitated (IP) with anti-H3 antibodies and subjected to Western (WB) analysis with anti-H3 and anti-Myc antibodies. In E, relative binding was calculated as the ratio of Myc pellet to supernatant signal divided by the H3 pellet signal (n = 3). F, ChIP analysis of Myc-Eaf1 at GAL1. WT, bre1Δ, dot1Δ, and gcn4Δ strains (DGY3, DGY427, DGY56, and DGY8) were grown in SCRaf at 30 °C, treated with 2% galactose for 30 min, and ChIP was performed using anti-Myc antibodies and PCR primers to amplify the GAL1 UAS or 3′ ORF and chromosome V in the presence of [α-33P]dATP. Occupancy was calculated, taking the ratio of radioactivities of PCR products for GAL1 versus the chromosome V reference and dividing by the same ratio for input samples. Occupancy in galactose was divided by the occupancy in raffinose.