FIGURE 4.
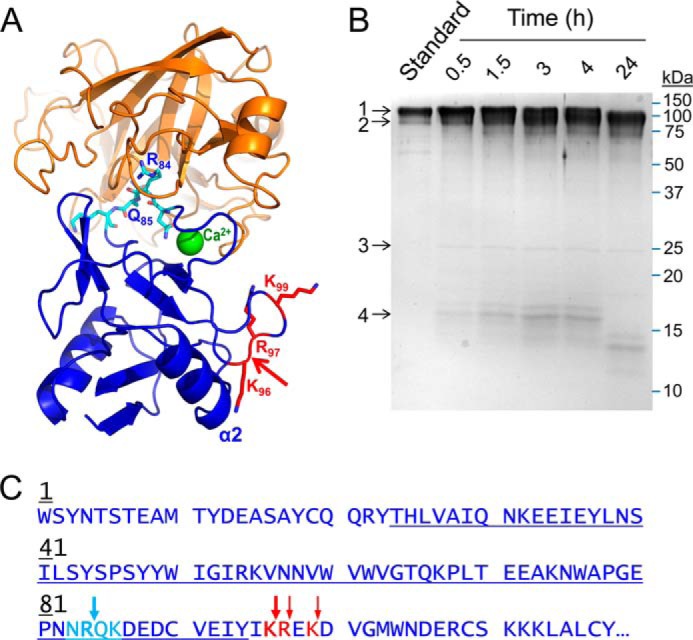
Proteolysis of recombinant human E-selectin by mesotrypsin. A, the crystal structure of mesotrypsin (orange; PDB entry 2R9P, chain B) docked with the C-type lectin domain of human E-selectin (blue; PDB entry 1ESL) in which the lectin residues 83–86 (cyan), which feature a canonical-like backbone conformation, are positioned in the mesotrypsin active site cleft. The molecular surfaces are roughly sterically compatible. The actual mesotrypsin cleavage sites in E-selectin that were subsequently identified experimentally are shown in red, and the primary initial nick site is indicated by the red arrow. In the absence of major conformational changes in E-selectin, binding of mesotrypsin to this cleavage site would be sterically blocked by the adjacent α2 helix. B, Coomassie-stained 15% SDS-polyacrylamide gel shows gradual cleavage of 5 μm recombinant human E-selectin (band 1) by 0.25 μm mesotrypsin (band 3) over a 24-h time course to generate a large C-terminal fragment (band 2) and a smaller N-terminal fragment (band 4), which was subsequently further degraded. The first lane shows a 1-μg undigested E-selectin run as a standard; subsequent lanes show time point samples collected at times indicated above the gel. C, the amino acid sequence of the C-type lectin domain of human E-selectin is shown; this domain encompasses residues 1–118 of the 589-residue mature secreted glycoprotein. The loop of canonical-like conformation is colored cyan, with the Arg84-Gln85 bond hypothesized to be a potential mesotrypsin cleavage site indicated by a cyan arrow. The actual mesotrypsin cleavage sites identified by N-terminal sequencing of the large cleavage fragment are indicated by red arrows. The blue underlining indicates the sequence coverage obtained from in-gel chymotrypsin digestion and nanoLC-MS/MS analysis of the 16-kDa cleavage fragment.
