Abstract
In this issue of Cell Stem Cell, Liu and co-workers demonstrate that a genetically engineered glioma model displays a functional cellular hierarchy defined by expression of the nuclear orphan receptor, Tlx. Targeting cancer stem cells through genetic deletion of TLX promotes cancer stem cell death and differentiation and extends survival.
Cancer is a genetic disease, but it initiates and grows within a cellular context that reflects the organization of tumors into aberrant organ systems. The cellular diversity within a tumor extends to the neoplastic compartment in which genetic and epigenetic variation manifests in differential proliferation, survival, and migration of cancer cells. The cancer stem cell (CSC) hypothesis is a partial explanation for these observations as many cancers harbor a relatively undifferentiated pool of transformed cells that self-renew and propagate the entire range of differentiated tumor progeny. However, establishing the presence of cellular hierarchies within a tumor and identifying the origin of differentiated progeny is complicated by the dynamic nature of cancer and an inability to trace the history of existing cancers. One challenge has been the identification of reliable CSC surface markers because these molecules mediate interactions between cells and their microenvironment and may be dynamically changed after isolation of CSCs. Further, no tumor type is genetically uniform so markers are unlikely to be uniformly informative in all tumors. Tumor models derived from patient specimens (e.g. patient-derived xenografts) contain the cellular and molecular diversity of the human disease, but cannot instruct us as to the natural history of specific cells beyond genetic lineage analysis. Alternatively, genetically engineered mouse models present a useful tool to study the natural history of tumor hierarchies in an immune competent background, albeit with far less genetic diversity than human cancers. Based on this background, Liu and colleagues (Zhu et al., 2014) established a mouse model that developed high-grade brain tumors by co-expression of PDGFB and AKT in Nestin positive neural stem cells (NSCs). Other investigators have used Nestin to mark highly tumorigenic cells, but the marker has not proven reliable in human tumors. In contrast, the current study interrogated Tailless (Tlx; Nuclear Receptor subfamily 2 group E, NR2E1), a nuclear orphan receptor specifically expressed in the brain and retina, to perform lineage tracing and targeting studies.
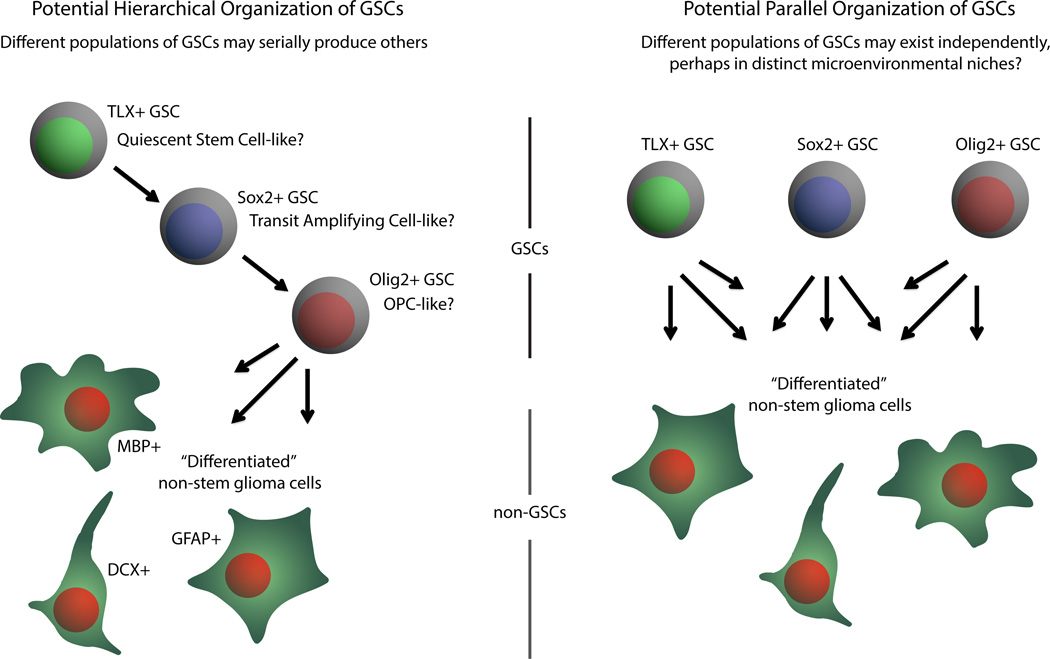
Previous studies by this group and others have shown that Tlx is expressed in neural stem cells (NSCs) (Liu et al., 2008; Shi et al., 2004) and that forced overexpression of Tlx in conjunction with common genetic lesions (e.g. mutant p53) induces gliomas (Liu et al., 2010; Park et al., 2010; Zou et al., 2012). Building on these findings, the authors used TLX-GFP reporter mice in conjunction with PDGFB and AKT overexpression to generate gliomas with variable GFP expression. TLX-GFP+ cells are largely quiescent and increased sphere formation and tumor propagation potential relative to TLX-GFP- cells. Notably, TLX-GFP+ cells were distinct from cells expressing other putative CSC markers (Sox2, Olig2), suggesting that tumors may contain different pools of CSCs and/or a further hierarchy of stem and progenitor cells (Figure 1). As both NSCs and oligodendroglial progenitor cells (OPCs) have been shown to be potential cells-of-origin for gliomas, it is possible that tumor growth in these genetic models may not be solely driven by a single cell type. Lineage tracing studies with Confetti reporter mice suggested that TLX+ cells can be the cell-of-origin of gliomas and that treatment with the oral methylator, temozolomide, promotes cell cycle entry of TLX+ cells. These results suggest that TLX is a potential molecular target of gliomas. Indeed, conditional ablation of TLX in their model slows tumor growth and inhibits CSC self-renewal, associated with induction of senescence and neurogenic differentiation measured by DCX expression. Interestingly, we previously reported DCX as a negative prognostic indicator for glioblastoma in combination with other genes (Rich et al., 2005).
Figure 1. The role of the nuclear orphan receptor TLX (NR2E1) in glioblastoma.
TLX-positive glioma cells are quiescent and display the capacity for self-renewal and tumor growth. TLX marks a subpopulation of tumor cells distinct from other putative cancer stem cell (CSC) markers – SOX2 and OLIG2 – suggesting the potential for different pools of CSCs or further stages in a cellular hierarchy with SOX2 potentially indicating a transient amplifying progenitor population
To further determine potential molecular mediators of TLX, the authors performed an expression analysis and found cell cycle regulators (including CDKN2A, CDKN2B, and PML) and neuronal differentiation genes (TGFβR1 and Dlx2) were significantly up-regulated in TLX knockout CSCs. Other potential molecules previously found to interact with TLX not detected in these studies could also be relevant based on roles in glioma CSCs, including Pten, miR-9, and LSD1. These findings are consistent with prior reports that TLX functions as a transcription repressor in controlling CSCs. TLX recruits histone deacetylases (including HDAC3 and HDAC5) to its downstream targets to repress their transcription (Sun et al., 2007). As HDACs are required for function of TLX transcriptional repressor and essential in the maintenance of CSCs self-renewal, HDAC inhibitors may target TLX+ CSCs.
The identification of novel and specific CSC targets is potentially important as CSCs contribute to therapeutic resistance (Bao et al., 2006). In support of TLX as a CSC target, the authors performed online in silico analysis using The Cancer Genome Atlas (TCGA) bioinformatics dataset to show that high TLX mRNA expression is a negative prognostic factor for an unselected glioblastoma population (P < 0.007, Cox Proportional Hazard). This observation must be taken with caution as a further examination of the full TCGA dataset with consideration of other prognostic factors indicates that the prognostic significance of TLX is entirely linked to its reduced expression in G-CIMP (glioma CpG island methylator phenotype) tumors, which signify a genetically distinct cancer type and are associated with IDH1 mutations and longer survival. Excluding G-CIMP patients, TLX expression displays no predictive value for glioblastoma patient survival (P = 0.955, Cox Proportional Hazard). Thus, TLX is not likely a prognostic factor itself, although the reduced TLX expression found in G-CIMP patients may potentially inform the biology of this distinct population of tumors. Additionally, genetic lesions (Pten, p53, etc.) potentially interacting with TLX may also inform its contribution to tumor growth. While no TLX inhibitors have been identified, the TLX mutant mouse is viable, albeit with developmental abnormalities in the brain, and TLX has been shown to be a druggable target (Benod et al., 2014).
The studies from Liu and colleagues (Zhu et al., 2014) lend further support to the importance of CSCs, while supporting TLX as a novel glioma CSC marker and expanding opportunities to investigate regulators of CSCs in a genetic model. The combined use of this powerful model with well characterized human tumor models should inform the discovery of other CSC points of fragility and could provide a useful tool to detect the initiating stages of brain cancer. Although the CSC hypothesis does not comprehensively explain all of tumor biology, CSCs as roots of many cancers represent an added level of complexity in tumors, a challenge we must face in trying to develop more effective therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006 Dec 7;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Benod C, Villagomez R, Filgueira CS, Hwang PK, Leonard PG, Poncet-Montange G, Rajagopalan S, Fletterick RJ, Gustafsson JÅ, Webb P. The Human Orphan Nuclear Receptor Tailless (TLX, NR2E1) Is Druggable. PLoS One. 2014 Jun 17;9(6):e99440. doi: 10.1371/journal.pone.0099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Belz T, Bock D, Takacs A, Wu H, Lichter P, Chai M, Schutz G. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes & development. 2008;22:2473–2478. doi: 10.1101/gad.479308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Wang Y, Belz T, Bock D, Takacs A, Radlwimmer B, Barbus S, Reifenberger G, Lichter P, Schütz G. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev. 2010 Apr 1;24(7):683–695. doi: 10.1101/gad.560310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim JK, Jeon HM, Oh SY, Kim SH, Nam DH, Kim H. The neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol Cells. 2010 Nov;30(5):403–408. doi: 10.1007/s10059-010-0122-z. [DOI] [PubMed] [Google Scholar]
- Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005 May 15;65(10):4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Khan MA, Weiler M, Blaes J, Jestaedt L, Geibert M, Zou P, Gronych J, Bernhardt O, Korshunov A, Bugner V, Lichter P, Radlwimmer B, Heiland S, Bendszus M, Wick W, Liu HK. Targeting Self-Renewal in High-Grade Brain Tumors Leads to Loss of Brain Tumor Stem Cells and Prolonged Survival. Cell Stem Cell. 2014 May 14; doi: 10.1016/j.stem.2014.04.007. pii: S1934-5909(14)00144-1. [DOI] [PubMed] [Google Scholar]
- Zou Y, Niu W, Qin S, Downes M, Burns DK, Zhang CL. The nuclear receptor TLX is required for gliomagenesis within the adult neurogenic niche. Mol Cell Biol. 2012 Dec;32(23):4811–4820. doi: 10.1128/MCB.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]