Abstract
Objective:
To test the feasibility of DNA barcoding for accurate identification of Jinqian Baihua She and its adulterants.
Materials and Methods:
Standard cytochrome C oxidase subunit I (COI) gene fragments were sequenced for DNA barcoding of 39 samples from 9 snake species, including Bungarus multicinctus, the officially recognized origin animal by Chinese Pharmacopoeia, and other 8 adulterate species. The aligned sequences, 658 base pairs in length, were analyzed for divergence using the Kimura-2-parameter (K2P) distance model with MEGA5.0.
Results:
The mean intraspecific K2P distance was 0.0103 and the average interspecific genetic distance was 0.2178 in B. multicinctus, far greater than the minimal interspecific genetic distance of 0.027 recommended for species identification. A neighbor-joining (NJ) tree was constructed, in which each species formed a monophyletic clade with bootstrap supports of 100%. All the data were submitted to Barcode of Life Data system version 3.0 (BOLD, http://www.barcodinglife.org) under the project title “DNA barcoding Bungarus multicinctus and its adulterants”. Ten samples of commercially available crude drugs of JBS were identified using the identification engine provided by BOLD. All the samples were clearly identified at the species level, among which five were found to be the adulterants and identified as Dinodon rufozonatum.
Conclusion:
DNA barcoding using the standard COI gene fragments provides an effective and accurate means for JBS identification and authentication.
Keywords: Bungarus multicinctus, cytochrome C oxidase subunit I, DNA barcodes, identification
INTRODUCTION
Jinqian Baihua She (JBS) (coin-like white-banded snake, Bungarus parvus) is a commonly used high-value traditional Chinese drug derived from the dried body (with the viscera discarded) of many-banded krait, Bungarus multicinctus multicinctus Blyth (Fam. Elapidae). According to Chinese Pharmacopeia,[1] JBS is effective in dispelling the wind, removing obstruction of the collaterals and relieving spasm. B. multicinctus has long been used as a folk medicine in its natural range, i.e. the south part of China, especially the tropical and subtropical mountainous regions with high humidity almost all through the year, as well as the regions with hot summer and cold winter such as the areas around Nanling Mountains (i.e. Guangdong, Guangxi, Hunan, Jiangxi, etc.). The residents in these regions often suffer from bone pain, arthritis, rheumatism, and even paralysis, and they improvised to regularly take self-prepared strong alcoholic drinks (in which the snake was immersed for preservation for at least months to allow the pharmaceutically active ingredients to be slowly released) to treat or prevent these symptoms.
Conventionally, JBS derived from the infant snakes (as small as a coin when coiled, hence the name “coin-like”) are thought to have stronger pharmacological actions. As the attempts of domestication and breeding of B. multicinctus currently prove unsuccessful, the supply of JBS depends entirely on the gradually diminishing wild resource. This supply shortage but huge demand of JBS results in its high price in market, and also in the emergence of adulterants, such as those using baby Dinodon rufozonatum, B. fascitus, etc., So far nine species of snakes have been found to be sold under the name of JBS.[2,3] The differentiation of these adulterants from the genuine JBS merely by their morphological characters is rather difficult because of their high similarities in appearance, especially for the young snakes [Figure 1]. The accurate identification depends heavily on the inspector's professional experience. To ensure the effectiveness and safety of the drug, it is necessary to find a convenient and accurate means for distinguishing genuine JBS from its adulterants.
Figure 1.
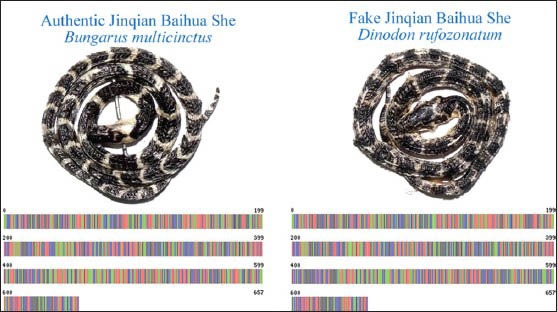
Authentic Jinqian Baihua She and one of its adulterants derived from Dinodon rufozonatum, and their COI barcodes
DNA barcoding, first proposed in 2003 by researchers at the University of Guelph in Ontario, Canada,[4] is a taxonomic method that uses a short genetic marker in an organism's DNA to identify the particular species of the organism. The short genetic sequence from a standard part of the genome works efficiently to identify the species in the way a supermarket scanner distinguishes products using the black stripes of the Universal Product Code; thus it was given the name “DNA barcode”. DNA barcodes vary among individuals of the same species, but only to a very minor degree. Normally, the minor variation of the DNA barcode region within species is trivial compared to the differences between species.
DNA barcode sequences are very short relative to the entire genome and can be obtained quickly and cheaply. The sequence of mitochondrial cytochrome C oxidase subunit 1 (COI) can serve as the standard barcode for almost all animals.[5] The COI sequence contains 648 base pairs in most groups, very short compared to the 3 billion base pairs in a higher animal genome. COI has proved to be highly effective in identifying birds,[6] butterflies,[7] fishes,[8,9] flies[10] and many other animal groups. Recently, new attempts at DNA barcoding of reptiles and snakes also yielded successful results. Using a newly developed set of reptile-specific primers for COI, Nagy et al. barcoded more than 250 species of reptiles in Madagascar, the world's fourth-largest island and a biodiversity hotspot.[11] Dubey et al. designed two sets of novel primers for targeting regions within the COI gene to produce 175 bp and 245 bp amplicons, and employed them as DNA mini-barcodes to identify some endangered snake species of India.[12]
As to JBS, Cui et al. made the first attempt on molecular identification of JBS based on COI barcode sequence.[13] However, they took only 4 species – B. multicinctus, B. fasciatus, Elaphe moellendorffi and Enhydris plumbea, with a total of 11 specimens – into consideration. Although it preliminarily proved the feasibility of DNA barcodes identification of JBS and its adulterants, the dataset was too small, and the inadequate data decreased its practicability. For instance, out of the 10 JBS samples we collected from the market, five failed to yield sensible results using Cui's database. A more comprehensive DNA barcode system was therefore needed for JBS's authentication and identification.
In the present study, we collected 39 specimens of 9 snake species reported to be used in the name of JBS and sequenced their DNA barcode region (COI). All the sequences were uploaded to BOLD system and approved by CBOL (The Consortium for the Barcode of Life) as legitimate barcodes. Furthermore, we validated the DNA barcodes database by authenticating JBS crude drug samples.
MATERIALS AND METHODS
Samples
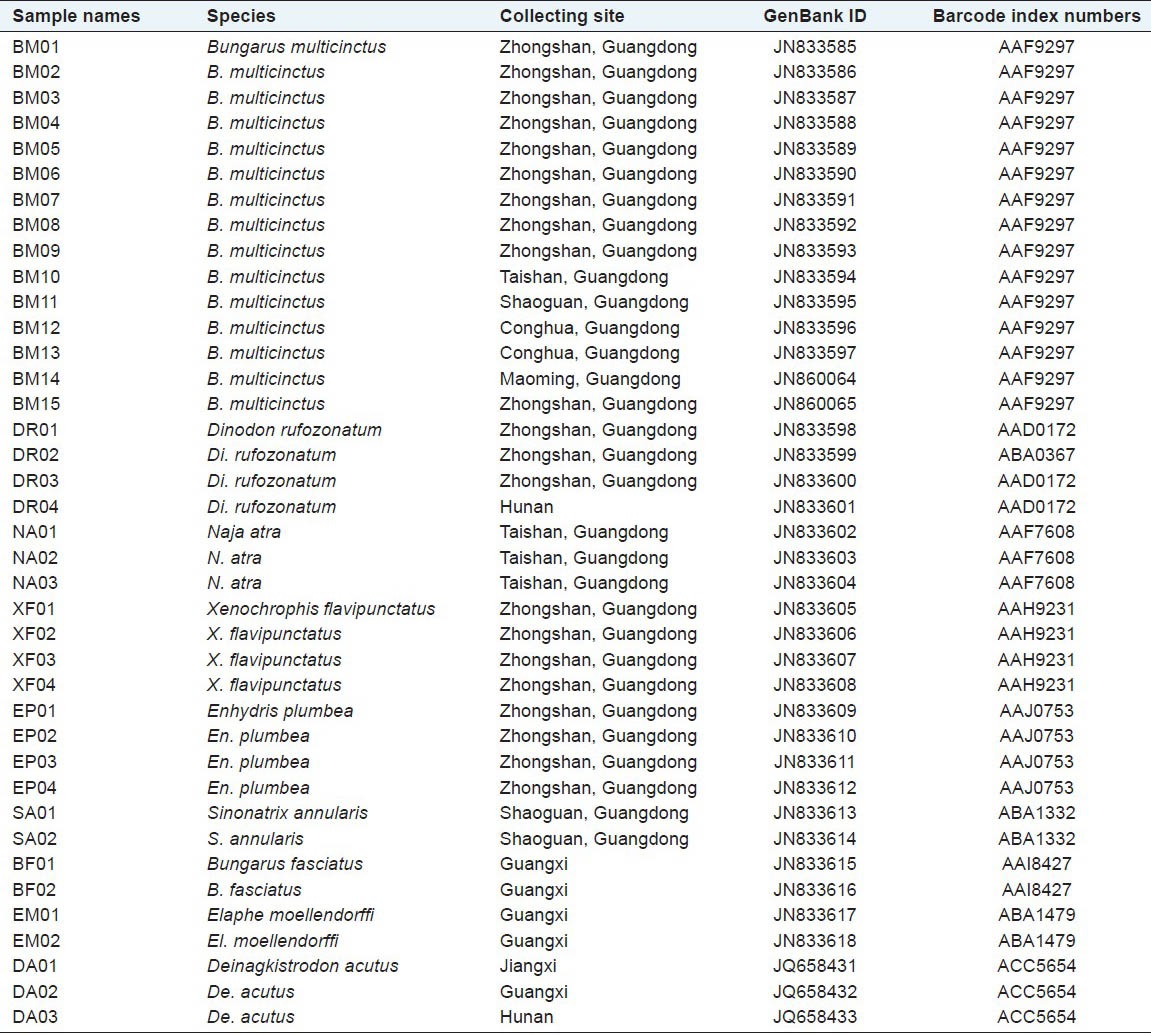
A total of 39 specimens from 9 snake species (including B. multicinctus and its adulterants) were obtained from various locations in Guangdong Province, Hunan Province, Jinagxi Province and Guangxi Zhuang Autonomous Region in China [Table 1]. Vouchers were deposited in School of Traditional Chinese Medicine, Southern Medical University, and all the specimens were preserved in 75% ethanol. Ten samples of JBS were purchased from the local drug stores or the crude drug market in Guangzhou, Guangdong Province.
Table 1.
Species and collecting sites of the specimens in this study
DNA extraction and PCR amplification
Tissue samples were dissected from the dorsal muscle of the snake. DNA was extracted using TIANamp Genomic DNA kit (Tiangen Biotech Co. Ltd., Beijing) following the manufacturer's instructions and was then dissolved in 100 μl distilled water.
COI barcode region was amplified using primers LCO1490 and HCO2198[14] in a total reaction volume of 25 μl containing 12.5 μl of 2 × Taq PCR Colorless Mix, 1 μl of each primer, 1 μl of genomic DNA, and 9.5 μl of ddH2O.
Thermal cycling was performed with an initial step at 93°C for 5 min and 55°C for 2 min, followed by 35 cycles of 93°C for 30 s, 55°C for 45 s, and 70°C for 45 s, with a final extension at 70°C for 5 min and chilling to 4°C. The PCR products were detected by 1.2% agarose gel electrophoresis, and visualized under ultraviolet light. After purification, the products were sequenced in both directions by Invitrogen Biotechnology (Shanghai) Co., Ltd.
DNA sequencing and COI barcode analysis
The DNA sequences were manually edited and aligned using the software BioEdit and Clustal X.[15] All the new data were submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and the Barcode of Life Database (BOLD, http://www.barcodinglife.org). The sequence divergences were calculated using the Kimura two-parameter (K2P) distance model,[16] and the genetic distances were computed by MEGA 5.0 software.[17] A bootstrap (1000 replicates) neighbor-joining (NJ) tree was constructed based on the K2P distances, to provide a graphical representation of the divergence patterns among the species. The NJ tree was then confirmed by bootstrapping to assign confidence levels to each branch in the tree.
Identification of JBS samples by BOLD identification engine
The 10 samples of JBS purchased from the local drug stores and crude drug market of Guangzhou were analyzed for COI barcode region sequences, and their species were identified with the identification engine provided by BOLD. On the other hand, we also invited an expert (the 5th author, Dr. Zhang L.) for professional inspection based upon morphological characters to verify the results produced by BOLD.
RESULTS
Sequence analysis of COI
The COI barcode regions of the 39 specimens representing 9 snake species under the name of JBS were sequenced. With an aligned length of 658 nucleotides, the sequences obtained contained no insertions, deletions, or codons. The nucleotide composition and variable sites were analyzed using MEGA 5.0. In total, 255 variable sites and 403 conserved sites were found in the COI sequences of B. multicinctus and its adulterants. The GC contents ranged from 40.1% to 47.4%, with an average of 44.3%; in the specific case of B. multicinctus, the GC content range was 42.9-43.3%.
Intraspecific variation and interspecific variation
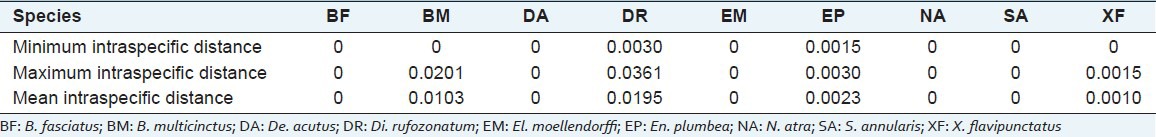
Altogether 14 variation sites were found in 15 B. multicinctus COI barcode sequences; the transition/transversion value was 6. The mean intraspecific K2P distance in B. multicinctus was 0.0103, with a maximum of 0.0201.
The intraspecific K2P distance of Di. rufozonatum, El. plumbea, and X. flavipunctatus ranged from 0.0030 to 0.0361, 0.0015 to 0.0030, and 0 to 0.0015, with the mean values of 0.0195, 0.0023, and 0.0010, respectively. Those of the other 5 species were all 0 [Table 2].
Table 2.
Intraspecific genetic distances of the 9 snake species
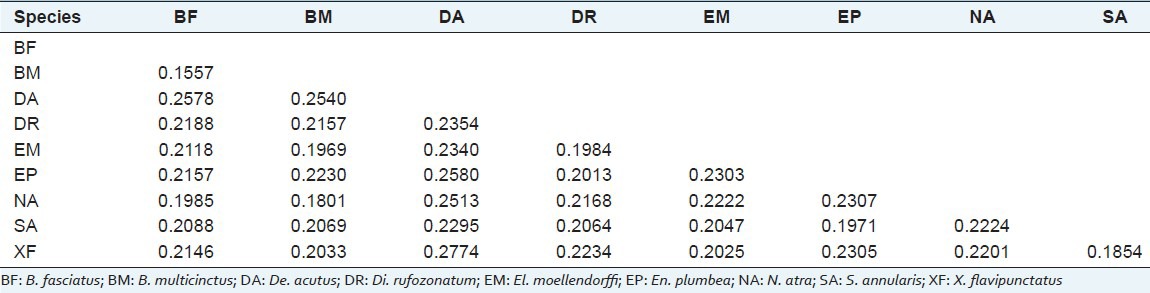
The average interspecific genetic distance was 0.2178. The minimum interspecific mean distance was found between B. multicinctus and B. fasciatus, which was 0.1557, far greater than that of 0.027 recommended by Hebert for species identification[4] (Hebert et al., 2003a). The maximum interspecific mean distance, 0.2578, was found between B. fasciatus and De. acutus [Table 3].
Table 3.
Mean distances between species
Neighbor-Joining tree
Neighbor-joining analysis was conducted to display the relationships between the analyzed species. In the NJ tree [Figure 2], each species formed a monophyletic clade with bootstrap supports of 100%. B. multicinctus could be differentiated clearly from the adulterants. It first clustered with B. fasciatus, a species of the same genus, and then with N. naja. These 3 species formed a branch representing Family Elapidae. The clades of Family Colubridae genera paralleled to Elapidae branch and combined with it to form a cluster joining Deinagkistrodon, Family Viperidae. Inside the clade of B. multicinctus, two subclades could be obviously recognized.
Figure 2.
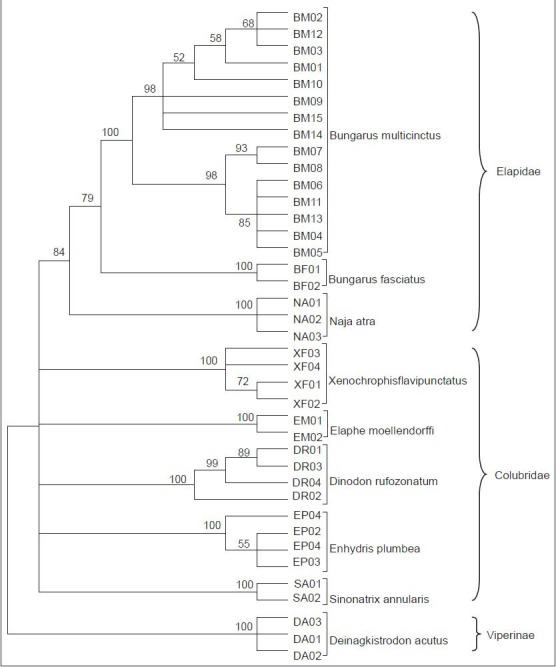
Neighbor-joining tree of 39 COI sequences from B. multicinctus and its adulterants (1,000 bootstrap replicates, Model = Kimura-2-Parameter)
JBS crude drug identification by BOLD
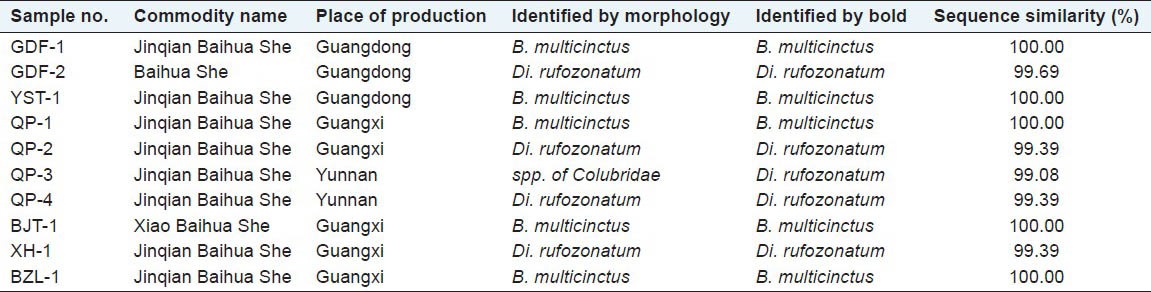
All of the 10 purchased crude drug samples of JBS were clearly identified at the species level with the BOLD identification engine [Figure 3]. Each sample gave a maximum identity of 99.08-100% in COI sequence to the matched species. The results were consistent with the opinions of the invited expert who examined the samples on a morphological basis [Table 4].
Figure 3.
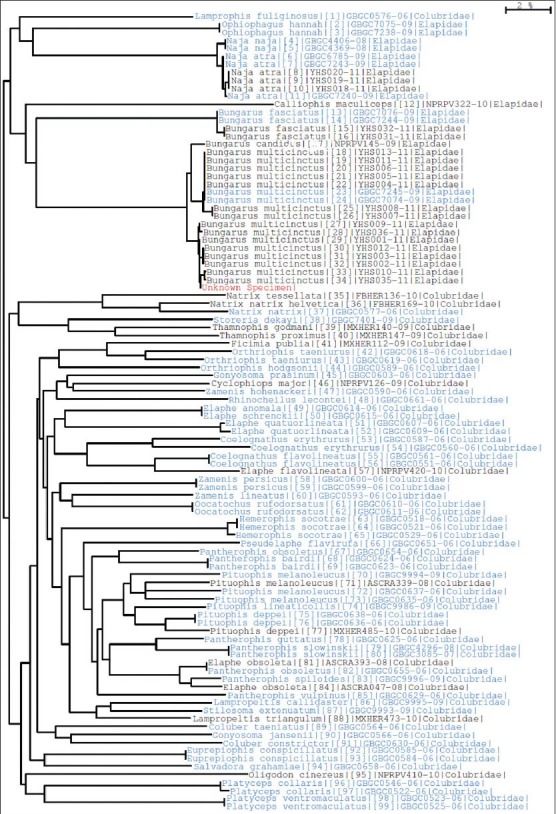
Identification result of a JBS crude drug sample YST-1 with BOLD identification engine. The sample, shown as “unknown specimen” in red, was clearly identified at the species level as B. multicinctus. 2% represents the scale length
Table 4.
Identification results of JBS crude drug samples
DISCUSSION
Crude drugs of an animal origin comprise an important part in the huge treasure house of Traditional Chinese Medicine. According to statistics, about 1580 animal species are medicinally valued in China. Fifty-three kinds of animal-derived crude drugs can be found in Chinese Pharmacopoeia (ChP), which accounts for 9.6% of the total of 553 recorded items. JBS is one of the commonly used animal drugs, and its origin animal, the highly venomous snake B. multicinctus, has long been used for treating rheumatism and joint pains resulting from wet weather and its effectiveness is acknowledged in ChP. Currently the drug is widely used all over the country, which leads to a great demand for JBS.
But another problem arises. Because artificial breeding of B. multicinctus remains unsuccessful, JBS supply depends entirely on wild resources. The scale of the slaughter in recent decades has been so great that B. multicinctus population is gradually diminishing. The species was listed in China Red Data Book of Endangered Animals[18] and IUCN Red List of Threatened Species 2012. Along with the more and more intensive shortage in JBS supply, its price soars up.
The high price of JBS gives rise to various versions of adulterants. The common practice of adulterants is decoloring the body bands of other snakes with decolorants, coloring the bands into white with paint, or even painting the bands on a snake without bands. Another practice is splitting the body of large genuine snakes into small strips and assembling them with the heads of other species to make several fakes. These false JBS are highly similar in appearance with the genuine ones, thus making morphology-based identification very difficult even for the professionals.
The development of molecular biological techniques provides a solution to the authentication of JBS. The DNA barcoding technique has proved effective in species identification, especially for animals. Its application in crude drug authentication and identification has developed rapidly in recent years and accelerated the standardization in identifying Traditional Chinese Materia Medica.[19] As to animal-derived crude drugs, the standard DNA barcode fragment, COI gene, has shown its power. Several successful instances of application of this technique included the differentiation of Cornu Cervi Pantotrichum (unossified antler of the spotted deer), Trionycis carapax (back shell of Chinese softshell turtle), Moschus (deer musk) and their adulterants, etc.[20,21,22]
Although Cui et al. preliminarily proved the feasibility of DNA barcodes identification of JBS and its adulterants,[13] the practicability of their work should be improved, as mentioned earlier in this paper. Investigation in a much larger scale was launched in the present study, and some new discoveries were made.
There are two primary criteria for an efficient DNA barcode: One is whether the individuals of a species have sequences of complete conformity in the candidate barcode region, the other is that the interspecies diversity must be much greater than the intraspecies variation.[6,7,23,24,25] We found 14 variable sites in the aligned 658-bp-long COI sequence, the standard DNA barcode region of B. multicinctus, whose mean and maximum intraspecies K2P distances were 0.0103 and 0.0201, respectively. The average genetic distance between the individuals of B. multicinctus and its adulterants was 0.2178, which was 10 times greater than the maximum K2P genetic distance within B. multicinctus. Moreover, in the cases of other species, the mean intraspecific K2P distances, ranging from 0 to 0.0195, were all far less than the minimum interspecific mean distance, 0.1557. Thus, we could say that COI well meet the criteria for use as an appropriate barcode for differentiating JBS and its adulterants.
Meanwhile, in a neighbor-joining tree constructed with 39 COI sequences from B. multicinctus and its adulterants, 9 clades with 100% bootstraP value could be recognized, each representing one species, including all the individuals from that species. These data demonstrate the high reliability of COI sequence in distinguishing B. multicinctus from its adulterants. When combined with Cui's data, we got a similar tree (data not shown).
We uploaded all the COI sequences to BOLD under the project title “DNA barcoding Bungarus multicinctus and its adulterants”. Using the identification engine provided by the system, we identified 10 crude drug samples of JBS collected from the local market. All the samples were clearly identified, including a sample that was unable to be recognized by the snake expert. This 100% success rate confirmed the accuracy of DNA barcoding in JBS authentication, and validated the database's practicability. What's more, we invited the pharmacy staff to identify the 10 JBS samples on a morphological basis. We found that most samples could not be authenticated by them. It shows that DNA barcode as a simple and efficient technique won’t be influenced by the professional level of researcher.
Based on our identification results shown in Table 4, it is estimated that about 50% of JBS sold in market was faked with Di. rufozonatum. So far, artificial breeding has succeeded with several snake species, including Di. rufozonatum, which can therefore be commercially available in large quantity and at a rather low price to make counterfeiting JBS a very profitable business. The high proportion of adulterants in JBS market urges much more rigorous supervision of JBS quality.
The B. multicinctus clade in the neighbor-joining tree was clearly divided into two subclades, which was the evidence for intraspecific differentiation. In China, two subspecies of B. multicinctus were recorded: one was B. multicinctus multicinctus and the other was B. multicinctus wanghaotingi. The latter distributes only in the Yunnan Province, far from the sample collection sites in Guangdong. We checked all the specimens of B. multicinctus thoroughly, and found no morphological characteristics of subspecies B. multicinctus wanghaotingi. We therefore presume that the intraspecific differentiation may be attributed to some environmental factors, especially the residential altitude of different snake populations. The same phenomenon was also observed in other snake species.[26] Nevertheless, this does not affect the results of COI barcoding for distinguishing B. multicinctus from its adulterants.
DNA barcoding also provides a means for helping ameliorate the endangered situation of B. multicinctus. The species’ endangerment results from not only its poor fertility and the vulnerable environment under destruction by human activities, but also from an excessive poaching and trafficking. Illegal trade and uncontrolled hunting have threatened the survival of several endangered snake species, and many effective conservation actions depend on accurate species identification. Several successful examples had been reported recently of using DNA barcoding for authenticating bioproducts and monitoring illegal species exploitation.[12,27,28,29] Our barcodes data of B. multicinctus can also lend support to the protection of the species.
A precondition for accurate identification of species by the DNA barcoding technique is the constant enrichment and improvement of the barcode database. We have deposited 39 COI sequences in BOLD, and our ongoing efforts of adding new data from more B. multicinctus and its adulterants specimens, especially the newly found adulterants, will make DNA barcode-based JBS authentication more accurate and more practical.
Molecular identification of animal-originated crude drugs has already been adopted by Chinese Pharmacopeia. In particular, highly specific PCR identification of Zaocys and Agkistrodon, 2 crude drugs derived from the snake species Zaocys dhumnades and De. acutus, respectively, were recorded. We suggest that DNA barcoding be used as a standard method for identification of medicinal animals including snakes in the next edition of Chinese Pharmacopeia.
CONCLUSION
We collected specimens of B. multicinctus and its adulterants, and set up a DNA barcode database based on COI sequences. It can be used efficiently for JBS crude drug identification, and our results have demonstrated the effectiveness of the DNA barcoding technique in JBS identification and authentication practice.
ACKNOWLEDGMENT
The study was supported by an open fund from “DNA Barcoding Medicinial Animals in China” Program, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences (No. MADNA201004), and China National Innovation and Entrepreneurship Training Programs for Undergraduates (No. 201212121056).
Footnotes
Source of Support: The study was supported by an open fund from “DNA Barcoding Medicinial Animals in China” Program, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences (No. MADNA201004), and China National Innovation and Entrepreneurship Training Programs for Undergraduates (No. 201212121056)
Conflict of Interest: None declared.
REFERENCES
- 1.State Pharmacopoeia Committee. Beijing, China: Chemical Industry Press; 2010. 2010. Pharmacopoeia of the People's Republic of China; p. 204. [Google Scholar]
- 2.Wang YQ, Zhou KY, Xu LS, Xu GJ. Authentication of Bungarus Parvus and its adulterants by DNA molecular method using diagnostic primer. Chin Pharm J. 2000;9:61–6. [Google Scholar]
- 3.Feng CQ, Tang XJ, Huang LQ, Qian ZZ, Zhang J, Cui GH. High specific PCR identification of Bungarus multicinctus and its adulterants. Chin J Chin Mater Med. 2006;31:1050–3. [PubMed] [Google Scholar]
- 4.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–21. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: Cytochrome c oxidase subunit divergences among closely related species. Proc Biol Sci. 2003;270:S96–9. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–7. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. DNA barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci. 2005;360:1847–57. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, et al. Bernatchez. Identifying Canadian fresh water fishes through DNA barcodes. PLoS One. 2008;3:e2490. doi: 10.1371/journal.pone.0002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PD. DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proc Natl Acad Sci U S A. 2007;104:4967–72. doi: 10.1073/pnas.0700050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy ZT, Sonet G, Glaw F, Vences M. First Large-Scale DNA Barcoding Assessment of Reptiles in the Biodiversity Hotspot of Madagascar, Based on Newly Designed COI Primers. Plos One. 2012;7:e34506. doi: 10.1371/journal.pone.0034506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey B, Meganathan PR, Haque I. DNA mini-barcoding: An approach for forensic identification of some endangered Indian snake species. Forensic Sci Int Genet. 2011;5:181–4. doi: 10.1016/j.fsigen.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Cui LN, Du H, Zhang H. Molecular Identification of Bungarus Multicinctus and Its Adulterants Based on COI Barcode Sequence. World Sci Technol Mode Tradit Chin Med Mater Med. 2011;13:424–8. [Google Scholar]
- 14.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9. [PubMed] [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmouqin F, Hiqqins DG. The Clustal X windows interface: Flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Dudley J, Nei M, Kumar S. Mega4: Molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 18.Zhao EM. Beijing: Science Press; 1998. China Red Data Book of Endangered Animals: Amphibia and Reptilia. [Google Scholar]
- 19.Chen SL, Pang XH, Han JP, Yao H, Luo K. Identification System and Perspective for DNA Barcoding Traditional Chinese Materia Medica. World Sci Technol Mode Tradit Chin Med Mater Med. 2011;13:747–54. [Google Scholar]
- 20.Zhang R, Liu CS, Huang LQ, Wang XY, Cui GH, Dong S. Study on the Identification of Cornu Cervi Pantotrichum with DNA Barcoding. Chin Pharm J. 2011;46:263–6. [Google Scholar]
- 21.Du H, Cui LN, Zhang H, Yao H, Song JY, Chen SL. Molecular Identification of Trionyx Sinensis and Its Adulterants. World Sci Technol Mode Tradit Chin Med Mater Med. 2011;13:429–34. [Google Scholar]
- 22.Du H, Sun JM, Cui LN, Zhang H. Identification on DNA of Musk with COI Number and Its Fakes. Jilin J Tradit Chin Med. 2011;31:451–68. [Google Scholar]
- 23.Xiao JH, Xiao H, Huang DW. DNA barcoding: New approach of biological taxonomy. Acta Zool Sin. 2004;50:852–5. [Google Scholar]
- 24.Meyer CP, Paulay G. DNA Barcoding: Error Rates Based on Comprehensive Sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savolainen V, Cowan RS, Vogler AP, Roderick GK. Towards Writing the Encyclopedia of life: An Introduction to DNA barcoding. Philos Trans R Soc Lond B Biol Sci. 2005;360:1805–11. doi: 10.1098/rstb.2005.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo P, Malhotra A, Thorpe RS, Creer S, Pook CE. Comments on the systematic status of specimens belonging to the genus Viridovipera (Serpentes: Viperidae: Crotalinae) from Sichuan and Yunnan provinces of southwestern China, with a redescription of V. yunnanensis. Herpetol J. 2009;19:151–62. [Google Scholar]
- 27.Ribeiro AO, Caires RA, Mariguela TC, Pereira LH, Hanner R, Oliveira C. DNA barcodes identify marine fishes of São Paulo State, Brazil. Mol Ecol Resour. 2012;12:1012–20. doi: 10.1111/1755-0998.12007. [DOI] [PubMed] [Google Scholar]
- 28.Bitanyi S, Bjørnstad G, Ernest EM, Nesje M, Kusiluka LJ, Keyyu JD, et al. Species identification of Tanzanian antelopes using DNA barcoding. Mol Ecol Resour. 2011;11:442–9. doi: 10.1111/j.1755-0998.2011.02980.x. [DOI] [PubMed] [Google Scholar]
- 29.Yesson C, Bárcenas RT, Hernández HM, Ruiz-Maqueda ML, Prado A, Rodríguez VM, et al. DNA barcodes for Mexican Cactaceae, plants under pressure from wild collecting. Mol Ecol Resour. 2011;11:775–83. doi: 10.1111/j.1755-0998.2011.03009.x. [DOI] [PubMed] [Google Scholar]


