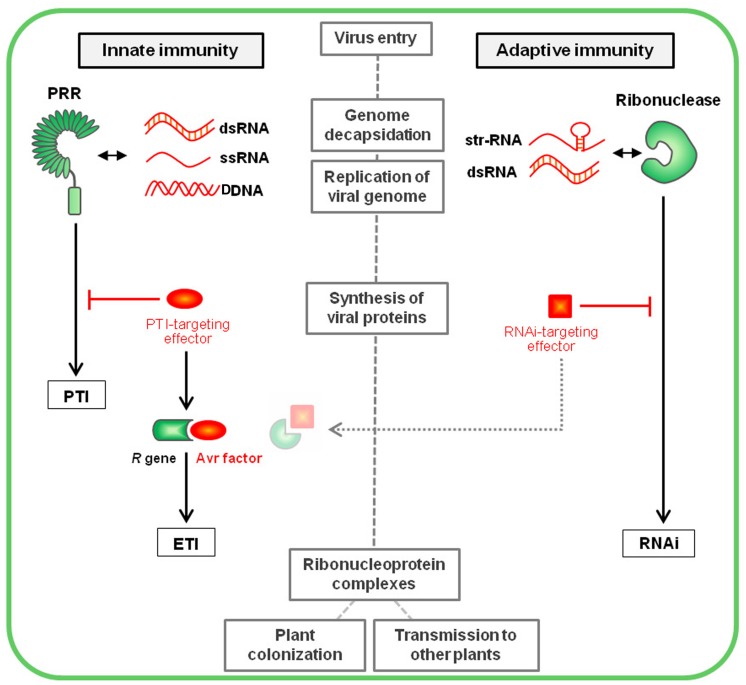
FIGURE 3.
Plant innate immunity vs. Plant adaptive immunity. Following entry into a host cell and genome decapsidation, the virus genome is liberated and replicated, leading to the accumulation of pathogenic nucleic acids that are perceived as viral PAMPs by specific intracellular PRRs. This recognition triggers a downstream cascade leading to PTI responses. Virus genome translation (occurring simultaneous to replication) leads to the synthesis of the virus-encoded proteins, among which viral pathogenic effectors enable the suppression of PTI signaling. In turn, specific plant R genes interact (directly or indirectly) with these effectors (that are then called avirulence factors) to trigger ETI. In the case of plant adaptive immunity illustrated by RNA interference (RNAi), the virus-derived elicitor molecules, corresponding to replicative dsRNAs or structured (str-RNA) viral genomes, are recognized by DCLs, key component of the silencing machinery that leads to virus degradation at the nucleic acid level. Viral proteins acting as RNAi-suppressing effectors interfere with this defense pathway. A recent publication (Sansregret et al., 2013) suggests silencing suppressors may be targeted by ETI-like mechanisms, restoring plant resistance. The main steps of the virus cycle into the first infected cell have been mentioned into gray boxes.