Figure 5.
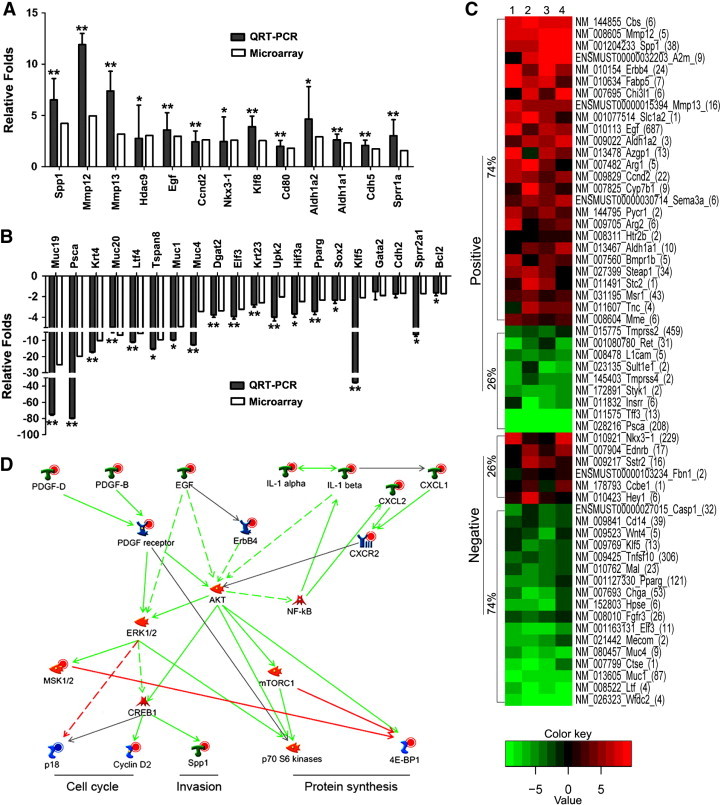
Bioinformatic identification and analysis of Klf5 deletion–dysregulated prostate cancer–related genes and molecular pathways in Pten-null mouse prostates. (A and B) Validation of differential expression for 13 upregulated (A) and 20 downregulated genes by real-time RT-PCR using the prostate samples including those used for microarray. White and black bars indicate microarray data and RT-PCR data, respectively. *P < .05; **P < .01. (C) Heat map of genes with > 2.0-fold expression change between wild-type and Klf5-null mouse prostates and associated with prostate cancer based on PubMed publications. The four wild-type prostates and the four Klf5-null prostates were randomly paired for map drawing. Genes are clustered on the basis of their association with prostate cancer, with those positively affecting prostate cancer cell behavior or upregulated in prostate cancer marked as “positive”, and those negatively affecting prostate cancer behavior or downregulated in prostate cancer marked as “negative”. The number in the parentheses after each gene name indicates the number of PubMed publications available on that gene. The percentage of upregulated or downregulated genes is shown. (D) Klf5 deletion activates canonical mitogenic signaling pathway involving AKT and ERK, as revealed by the analysis of Klf5 deletion–dysregulated genes using the MetaCore program. Up-regulation of signal initiating growth factors, cytokines, chemokines, and receptors activates downstream effectors involved in different processes through multiple canonical signaling pathways. Different shapes of the nodes represent functional classifications of genes. Red and blue circles in the nodes indicate up-regulation and down-regulation, respectively, of genes. Lines between every two nodes indicate their interactions, with red for inhibition, green for activation, gray for unspecified interaction, and dashed for indirect interaction. Arrow indicates the direction of a regulation.
