Figure 4.
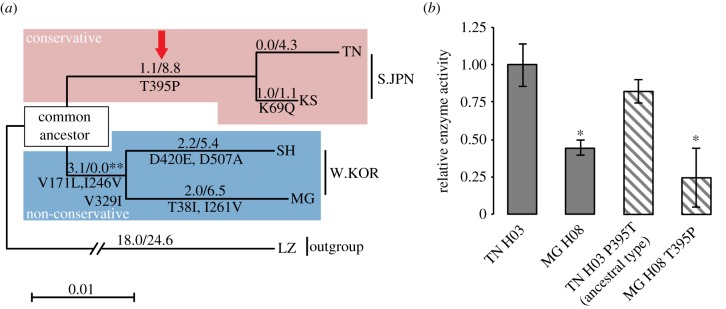
Reconstruction of ancestral CYP1B1. (a) Maximum-likelihood tree with synonymous and non-synonymous SNPs of Oryzias genus. LZ (O. lusonensis) from the Philippines was used as the out-group. The numbers on each branch show dN/dS (ω). Amino acid substitutions are indicated under each branch. The square on the tree indicates common ancestral sequence of S.JPN and W.KOR CYP1B1 inferred based on this tree; significant likelihood ratio test, **p < 0.02 (see also electronic supplementary material, table S10), are shown under the square. The red arrow shows the amino acids replaced in subsequent mutagenesis analyses. (b) Comparison of enzyme activities between wild-type and mutant enzymes. Constructs were generated using site-direct mutagenesis. This figure indicates that substitution of 395th amino acid had no effect on CYP1B1 enzyme activity. Maegok T395P was also generated to confirm the effect of the amino acid change. Each bar represents the mean ± s.d. from multiple independent samples (n = 3). Significant differences against Tanabe allele were identified using Games–Howell tests; *p < 0.05. (Online version in colour.)
