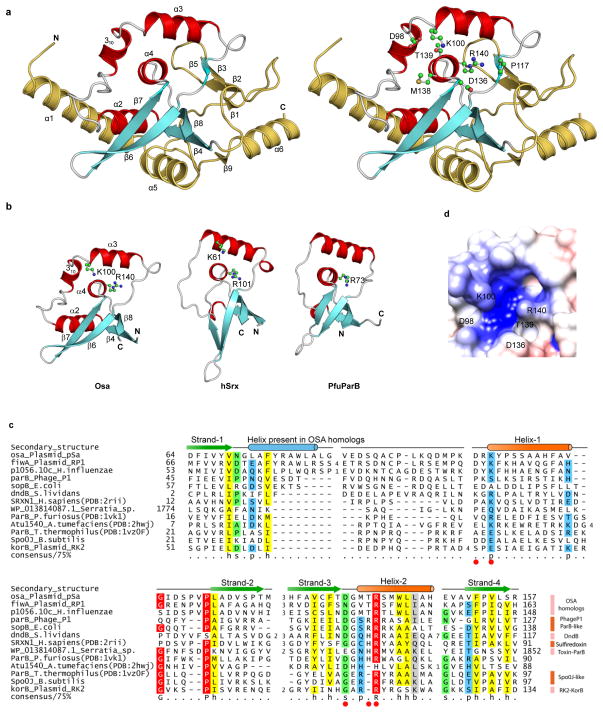
Figure 1.
Crystal structure of Osa with predicted active site residues. (a) Cartoon representation of Osa (left) showing the position of conserved residues (right). Helices and strands corresponding to the conserved structural core of the ParB/Sulfiredoxin fold are in red and cyan respectively. Additional lineage-specific structural elements are colored golden yellow. (b) Conserved structural core of the ParB/Sulfiredoxin fold: Osa (64–158), human Sulfiredoxin (hSrx:12–91, PDB:2rii) and ParB (Spo0J) from P. furiosus (PfuParB:16–90, PDB:1vk1). (c) Multiple sequence alignment of the ParB/Srx superfamily. Predicted active site residues are shown as red circles. Sequences are labeled by their gene and species names. The alignment is colored based on 75% consensus using the following scheme; p: polar residues (CDEHKNQRST) shaded blue, h: hydrophobic (ACFILMVWY) residues shaded yellow, s: small (ACDGNPSTV) residues shaded green, and big (QRKEILMWYF) residues shaded grey. Absolutely conserved residues are shaded red. Secondary structure is shown above the alignment with the core ParB/Srx strands in green and helices in orange. The blue helix is only conserved in Osa homologs. (d) Electrostatic surface representation of the predicted active site of Osa is shown with putative active site residues marked.