Figure 1.
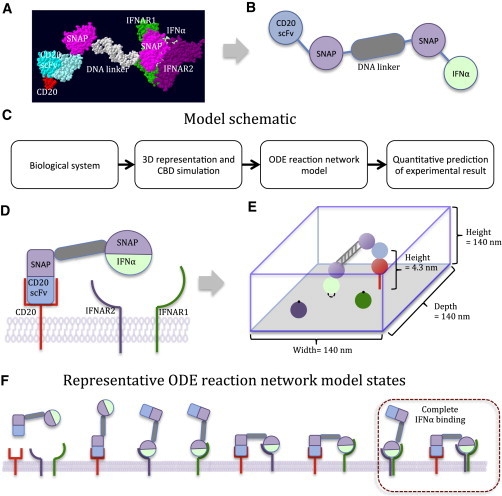
Two-component simulation model. (A) All-atom representation of the chimeric activator complex. The complex consists of an anti-CD20 scFv-SNAP fusion protein conjugated to an oligonucleotide that is hybridized to its complementary strand, which in turn is conjugated to an IFNα-SNAP fusion protein. (B) Coarse-grained representation of the chimeric activator complex. Each domain of the chimeric activator complex is represented as a simple geometric object for simulation. (C) Schematic of the CBD and ODE simulation model. (D) Simplified representation of complex with its binding partners, CD20, IFNAR1, and IFNAR2. (E) CBD simulation approach to computing the effect of the linker on targeted IFNα binding. We create a 3D representation of the complex with the anti-CD20 scFv bound to its antigen on the target (CD20+) cell surface. The model then tracks the constrained Brownian motions of the components of the complex and calculates the accessibility of IFNα to its receptors. (F) ODE model for tracking chimeric activator binding states. The ODE model simulates and tracks the kinetic transitions of all possible bound states of an anti-CD20 scFv-IFNα complex on the cell surface. When IFNα is bound to both IFNAR1 and IFNAR2, we refer to this state as complete IFNα binding, which leads to Stat1 phosphorylation. The separate entities in the diagram correspond to populations tracked in the ODE model. To see this figure in color, go online.
