Graphical Abstract
Keywords: Parasites, Zoonoses, Human impact, Captivity, Wildlife, Marine ecosystem
Highlights
-
•
We examine the presence of Giardia and Cryptosporidium in Australian sea lions.
-
•
The human associated Giardia duodenalis assemblages AI and B present in sea lions.
-
•
Proximity of sea lions to human settlements associated with occurrence of Giardia.
-
•
High presence of G. duodenalis in captive compared with wild sea lion populations.
-
•
Optimized PCR methodology required to determine transmission routes to sea lions.
Abstract
Giardia and Cryptosporidium are amongst the most common protozoan parasites identified as causing enteric disease in pinnipeds. A number of Giardia assemblages and Cryptosporidium species and genotypes are common in humans and terrestrial mammals and have also been identified in marine mammals. To investigate the occurrence of these parasites in an endangered marine mammal, the Australian sea lion (Neophoca cinerea), genomic DNA was extracted from faecal samples collected from wild populations (n = 271) in Southern and Western Australia and three Australian captive populations (n = 19). These were screened using PCR targeting the 18S rRNA of Giardia and Cryptosporidium. Giardia duodenalis was detected in 28 wild sea lions and in seven captive individuals. Successful sequencing of the 18S rRNA gene assigned 27 Giardia isolates to assemblage B and one to assemblage A, both assemblages commonly found in humans. Subsequent screening at the gdh and β-giardin loci resulted in amplification of only one of the 35 18S rRNA positive samples at the β-giardin locus. Sequencing at the β-giardin locus assigned the assemblage B 18S rRNA confirmed isolate to assemblage AI. The geographic distribution of sea lion populations sampled in relation to human settlements indicated that Giardia presence in sea lions was highest in populations less than 25 km from humans. Cryptosporidium was not detected by PCR screening in either wild colonies or captive sea lion populations. These data suggest that the presence of G. duodenalis in the endangered Australian sea lion is likely the result of dispersal from human sources. Multilocus molecular analyses are essential for the determination of G. duodenalis assemblages and subsequent inferences on transmission routes to endangered marine mammal populations.
1. Introduction
Protozoan parasites are a primary cause of morbidity and mortality in terrestrial and marine mammal populations (Hughes-Hanks et al., 2005; Ryan et al., 2014). Two genera, Cryptosporidium and Giardia, are amongst the most common organisms identified as causing enteric disease in pinniped species (Olson et al., 1997; Deng et al., 2000; Appelbee et al., 2005, 2010; Hughes-Hanks et al., 2005; Santin et al., 2005; Dixon et al., 2008; Bass et al., 2012). Currently, 26 Cryptosporidium species are considered valid and over 40 genotypes or cryptic species have been identified (c.f. Ryan et al., 2014). Several Cryptosporidium species present in humans and terrestrial mammals have been identified in marine mammals, including C. parvum, C. muris and C. hominis (Olson et al., 1997; Deng et al., 2000; Appelbee et al., 2005, 2010; Hughes-Hanks et al., 2005; Santin et al., 2005; Dixon et al., 2008). In addition Cryptosporidium seal genotypes 1 and 2, thought to be specific to marine mammals, have been identified in the ringed seal (Phoca hispida), and seal genotype 2 in the harbor seal (Phoca vitulina) and grey seal (Halichoerus grypus) (Santin et al., 2005; Bogomolni et al., 2008; Dixon et al., 2008). More recently, novel genotypes of Cryptosporidium have been described in the southern elephant seal (Mirounga leonina), harp seal (Pagophilus groenlandicus) and Weddell seal (Leptonychotes weddellii) (Rengifo-Herrera et al., 2011, 2013; Bass et al., 2012).
Of the six Giardia species, Giardia duodenalis has the broadest host range (Feng and Xiao, 2011). Molecular characterization of G. duodenalis has revealed significant genetic diversity, an indicator of complexity within this species. Accordingly, G. duodenalis is divided into assemblages A–H (c.f. Feng and Xiao, 2011), with assemblages A and B being the most diverse with at least four sub-assemblage types (I–IV) (c.f. Monis and Thompson, 2003). Assemblages A and B are human infective and have also been documented in wildlife and domestic animals (c.f. Feng and Xiao, 2011). Assemblages C and D are found in dogs, assemblages E, F and G in domestic ruminants, cats and rodents respectively, while assemblage H has been described in seals (c.f. Cacciò et al., 2005; Lasek-Nesselquist et al., 2010).
Infections of G. duodenalis are reported with a greater frequency in marine mammals than Cryptosporidium. Giardia assemblages A and B are the most commonly identified assemblages in pinniped species (Appelbee et al., 2005, 2010; Gaydos and Miller, 2008; Lasek-Nesselquist et al., 2008, 2010). Assemblages C and D have been described in harbor seals (Phoca vitulina) and the novel seal genotype H identified in grey seals (Gaydos and Miller, 2008; Lasek-Nesselquist et al., 2010). The presence of human host specific Giardia assemblages in marine mammals may be an indication of human impacts on the marine environment which poses potential concerns for conservation of endangered pinniped species.
The Australian sea lion (Neophoca cinerea) is one of the rarest seals in the world, with a total population of less than 15,000 (Shaughnessy et al., 2011). Colonies of this endangered pinniped are distributed on coastal islands and the mainland of Western and South Australia, many within close proximity to human settlements (Goldsworthy et al., 2007; Goldsworthy and Gales, 2008). As a tourist icon, populations on Kangaroo Island, South Australia, experience frequent human visitation and habitat disturbance. Interactions with, and proximity to, humans and wastewater run-off increases the likelihood of transmission of Giardia and Cryptosporidium from humans and domestic animals to seal populations.
The aim of this study was to detect and characterize Giardia and Cryptosporidium in wild and captive Australian sea lions within a phylogenetic framework, and to determine if proximity to human settlements was related to parasite detection. We hypothesized that if transmitted through human influences, protozoal strains would be more likely to be detected in seal colonies in close proximity to human settlements and would be similar to those found in domestic animals and human populations.
2. Materials and methods
2.1. Sample collection
Faecal samples from wild Australian sea lion colonies (n = 271) were opportunistically collected over a range of seasons during a 2 year period from 11 coastal and island colonies in Western Australia (Fig. 1A) and South Australia (Fig. 1B and C). Captive animal faecal samples (n = 19) were collected over a period of 4 months from the resident populations held at Dolphin Marine Magic and Taronga Zoo, New South Wales, and Sea World, Queensland. Faecal samples were transported to the laboratory and stored at 4 °C until processing for genomic DNA extraction.
Fig. 1.
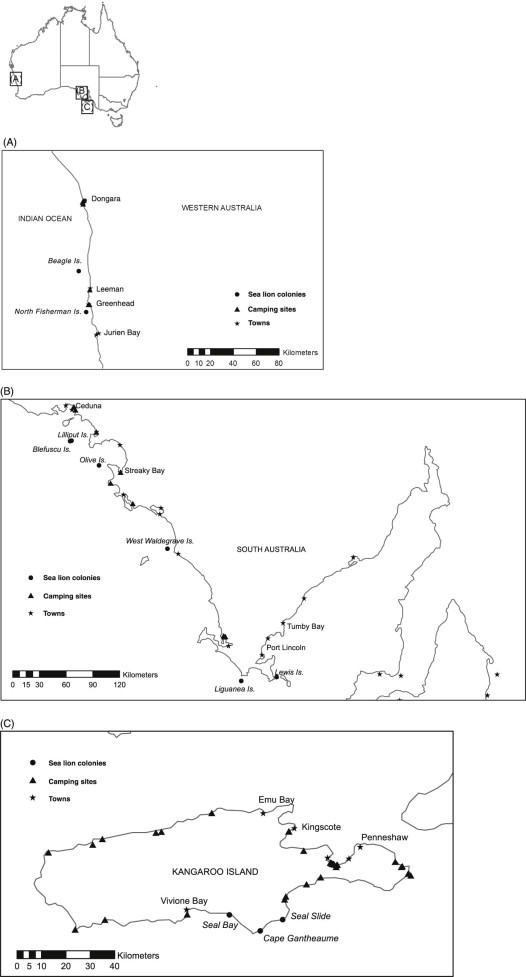
(A) Western Australia sampling locations. Faecal samples were collected from West Australia Sea lion colonies on Beagle and North Fisherman Islands. Coastal settlements and human impacted camping locations within close proximity to Sea lion colonies are indicated. (B) South Australia sampling locations. Australian sea lion faecal samples were collected from South Australia colonies; Blefuscu, Lewis, Liguanea, Lilliput, Olive and West Waldegrave Islands. Coastal towns and camping areas within close proximity to Australian Sea lion colonies are identified. (C) South Australia sampling locations: Kangaroo Island. Three colonies were sampled from Kangaroo Island including Cape Gantheaume, Seal Bay and Seal Slide. Coastal towns and recreational beach camping sites on the island are indicated.
2.2. DNA extraction
Genomic DNA was extracted from sea lion faecal samples (n = 290) using the ISOLATE Fecal DNA Kit (Bioline, Sydney, Australia). Faecal samples (approximately ~150 mg) were aliquoted into lysis bead tubes and DNA extraction performed as per the manufacturers protocol. Eluted DNA was stored at –20 °C until further analysis.
2.3. PCR screening for characterization of Giardia duodenalis
The presence of Giardia isolates was determined using the protocol targeting the 18S rRNA gene described in Hopkins et al. (1997) and Read et al. (2004). Nested PCR using the primers RH11/RH4LM in the primary reaction and GiAR18SeR/GiAR18SiR in the secondary reaction were used to amplify a ~ 175 bp fragment of the 18S rRNA gene. Primary and secondary reactions (25 µL) were prepared using the GC-RICH PCR system (Roche Diagnostics, Indianapolis, IN) (Asher et al., 2012). Thermocycling for both the primary and secondary reactions were performed using the conditions described by Hopkins et al. (1997).
To characterize Giardia assemblages 18S rRNA positive isolates (n = 35) were screened at the gdh and β-giardin loci. For gdh amplification the previously described semi-nested protocol of Read et al. (2004) was used. Primary and secondary reactions (25 µL) were prepared using the GC-RICH PCR system (Roche Diagnostics) and the primer set GdheF/GdhiR in the primary reaction and primers GdhiF/GdhiR in the secondary. Cycling was performed at denaturation for 2 min at 94 °C, 1 min at 56 °C, 2 min at 72 °C; 35 cycles at 94 °C for 30 s, 56 °C for 20 s and 72 °C for 45 s; and a final extension step at 72 °C for 7 min (Read et al., 2004). Amplification of the β-giardin locus was achieved following the nested protocol described by Cacciò et al. (2002) and Lalle et al. (2005). Primary reactions (25 µL) were prepared using 1.5 mM MgCl2, 200 µM dNTPs, 200 nM of each primer G7/G759, 1U of DNA polymerase Tth Plus (Fisher Biotec, Wembley, Australia) and 2 µL template DNA and the cycling conditions used by Cacciò et al. (2002). Secondary reactions (25 µL) were prepared using PCR chemistry identical to the primary reactions, with 2 µL of primary PCR product and following the thermocycling conditions described by Lalle et al. (2005). PCR was performed with the internal primers described by Lalle et al. (2005) with a slight modification to the internal forward primer (GAA CGA GAT CGA GGT CCG) after β-giardin sequence comparisons available on the NCBI GenBank database (http://ncbi.nlm.nih.gov/genbank/) showed a 4 bp difference between Giardia sequences and the internal forward primer sequence.
A spike analysis using DNA extracted from an existing Giardia laboratory trophozoite isolate was performed on 18S rRNA positive samples that failed to amplify at the gdh and β-giardin loci.
2.4. PCR screening for characterization of Cryptosporidium sp.
Screening for Cryptosporidium was conducted using a nested PCR protocol targeting the small subunit 18S rRNA (Xiao, 1999). RedHot Taq (Thermo Scientific, Scoresby, Australia) was used for reactions and all conditions were as described by Xiao et al. (2000). To confirm that the absence of Cryptosporidium was not the result of faecal inhibitors impairing DNA amplification, all samples (n = 290) were spiked with Cryptosporidium parvum DNA from an existing laboratory isolate (Waldron et al., 2011) and PCR screening repeated as described above.
All PCRs were performed in an Eppendorf Mastercycler (Eppendorf, North Ryde, Australia). PCR products (8 µL) were resolved by agarose gel electrophoresis (2% w/v, 110 V for 30 min) in TBE (Tris, boric acid, EDTA pH8) with 2 µL SYBR safe (Invitrogen, Mulgrave, Australia) using a HyperLadder II DNA marker (Bioline) to estimate amplicon size.
2.5. DNA sequencing and phylogenetic analyses
To identify Giardia assemblages the 18S rRNA products (n = 35) from GiAR18SeR/GiaR18SiR secondary reaction were purified for sequencing using the QIAquick PCR Purification Kit (Qiagen, Melbourne, Australia) and sequenced in the forward direction using the internal primer GiAR18SeR and in the reverse direction using GiAR18SiR. To identify Giardia sub-assemblage the β-giardin product (n = 1) was purified and sequenced in the forward and reverse direction using the secondary β-giardin PCR primers. All sequencing was performed by Macrogen Inc. (Seoul, Korea) on a 3130x1 Genetic Analyser (Applied Biosystems, Foster City, CA) using the standard run protocol for a 50 cm, 16 capillary array using a Big Dye terminator kit (Applied Biosystems).
Forward and reverse sequences (18S rRNA and β-giardin) were checked manually and trimmed in GeneiousPRO version 5.0.3 (Biomatters Ltd, Auckland, New Zealand) and a single contiguous sequence (contig) was assembled for each sample. BlastN sequence searches were performed to assign contiguous 18S rRNA and β-giardin sequences to an assemblage.
To allow for assemblage identification sequences were analysed within a phylogenetic framework. Representative 18S rRNA sequences for Giardia assemblages A–G were obtained from the NCBI GenBank database using accession numbers AF199446, AF199447, AF199449, AF199443, AF199448, AF199444, AF199450 for 18S rRNA analyses (Wielinga and Thompson, 2007). 18S rRNA and β-giardin sequences representing assemblage H were not available on GenBank and could not be included in analyses. Representative β-giardin sequences for Giardia assemblages AI–III to G were obtained from the NCBI GenBank database using accession numbers X85958, AY072724, FJ971410, AY072727, AY545646, AY545647, DQ116608, AY647264, EU769221 (Lalle et al., 2005; Wielinga and Thompson, 2007; Kosuwin et al., 2010; Lebbad et al., 2010). Contiguous 18S rRNA and β-giardin sequences generated in this study were aligned to GenBank sequences using ClustalW (Thompson et al., 1994) in MEGA version 6.0 (Tamura et al., 2013). For phylogenetic analyses, nucleotide substitution models were tested for maximum likelihood in MEGA6 (Tamura et al., 2013). Akaike Information Criterion corrected (AICc) values were used to determine the optimal parameters. Phylogenetic trees were constructed for 18S rRNA and β-giardin sequences using maximum likelihood (Tamura 3-parameter distance model with the uniform distribution parameter) and bootstrap analysis (1000 replicates) and compared with existing assemblages (Lalle et al., 2005; Wielinga and Thompson, 2007; Kosuwin et al., 2010; Lebbad et al., 2010). 18S rRNA sequences generated in this study have been submitted to the European Nucleotide Archive (ENA) under the accession numbers LN610171-LN610198. The β-giardin sequence generated in this study has been submitted to GenBank under accession number KM497498.
2.6. Mapping and statistical analyses
Maps illustrating the locations of wild sea lion populations sampled and proximity of towns and camping grounds were developed using ArcGIS version 10.0 (ESRI Inc, 2010).
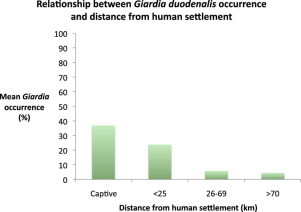
A Pearson's χ2 test was used to identify differences in the occurrence of Giardia duodenalis between wild and captive populations. For the wild populations only, a generalized linear model (GLM) with a binomial probability distribution was used to examine the effect of sea lion colony distance from human settlements and sampling season on presence/absence of Giardia. For this analysis, colonies were grouped into three distance-from-settlement categories: <25, 26–69 and >70 km (Table 1). Differences in occurrence between distance categories were determined using a Tukey's post-hoc test.
Table 1.
Australian sea lion colony groupings and analysis of Giardia duodenalis presence. Wild sea lion colony distance-from-settlement categories. Differences in occurrence between distance categories were determined using a Tukey's post-hoc test.
| Distance category (km) | Colonies in category | Mean occurrence of Giardia duodenalis (%) | Total number of samples |
|---|---|---|---|
| <25 | Beagle Island Cape Gantheaume North Fisherman Island Seal Bay Seal Slide |
23.8a | 80 |
| 26–69 | Blefuscu Island Lewis Island Liguanea Island Lilliput Island |
5.8a | 120 |
| >70 | Olive Island West Waldegrave Island |
2.8a | 71 |
Significant difference in Giardia duodenalis presence between groupings.
3. Results
3.1. Giardia detection and species identification
Screening of genomic DNA using a Giardia specific 18S rRNA protocol resulted in the detection of Giardia in 28 samples from wild sea lions (10.3%) and in seven samples from captive sea lions (36.8%). There was a significant difference in Giardia presence between wild and captive individuals (χ2 = 11.758, df = 1, p = < 0.001). In wild colonies, the distance from human settlement had a significant effect on the presence or absence of Giardia (Wald χ2 = 39.078, df = 2, p = < 0.001). Colonies less than 25 km from human settlements had a higher occurrence of Giardia than colonies more than 26 km away (Table 1). There was no effect of sampling season on Giardia presence (Wald χ2 = 6.112, df = 3, p = 0.106).
DNA sequences were obtained for 28 of the 35 18S rRNA positive samples. BlastN search identified 27 sequences as belonging to Giardia duodenalis assemblage B and one belonging to assemblage A. Analysis using the phyologentic framework clustered all samples from wild sea lions (n = 24) and three samples from captive animals within a clade that also contained the assemblage B reference sequence from GenBank (Fig. 2). One captive sample clustered within a clade containing the reference sequences from GenBank for assemblages A, E and F. Analysis of clustalW alignment showed a 2 bp polymorphism between the assemblages with the captive sample most closely aligned to assemblage A. Alignment of the sample to representative sequences showed that the sample was 100% identical to assemblage A but not E or F.
Fig. 2.
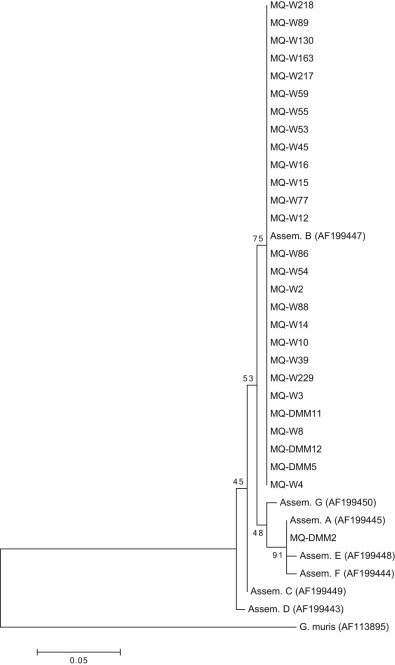
Giardia duodenalis 18S rRNA phylogenetic tree. Phylogenetic analysis of Giardia duodenalis positive samples was performed using a fragment of 18S rRNA gene. Analysis within the phylogenetic framework placed sea lion samples within the assemblage B (n = 27) and assemblage A clades (n = 1). Branch values indicate percent bootstrapping using 1000 replicates.
18S rRNA positive samples (n = 35) failed to amplify at the gdh locus. Representative samples all produced a gdh amplicon when spiked with Giardia isolate DNA. The β-giardin locus amplified in one of 35 18S rRNA positive samples identified as assemblage B. A DNA sequence was obtained for the β-giardin positive sample and a BlastN search identified the sequence as belonging to Giardia duodenalis assemblage AI. The inferred phylogeny placed the sample within a clade that also contained the assemblage AI reference sequence from GenBank. All samples spiked with Giardia positive DNA produced an amplicon when screened using β-giardin PCR.
3.2. Cryptosporidium screening
Cryptosporidium was not detected in any of the faecal samples (n = 290). The purified genomic DNA were deemed PCR competent using DNA spike analysis with all 290 samples generating an amplicon when screened using 18S rRNA PCR.
4. Discussion
In this study we examined the occurrence of protozoan parasites in the endangered Australian sea lion. Giardia duodenalis assemblage B, commonly found in humans and terrestrial mammals, was detected in wild and captive sea lion populations and G. duodenalis assemblage A was detected in a captive animal. Screening for Cryptosporidium failed to identify this parasite in any of the samples. Infections of Cryptosporidium are commonly reported at lower frequencies than Giardia in marine mammal populations (Appelbee et al., 2005, 2010; Gaydos and Miller, 2008; Lasek-Nesselquist et al., 2008, 2010; Hueffer et al., 2011). Of the eight Giardia duodenalis assemblages A and B are the most commonly identified in wild seal and sea lion populations (Appelbee et al., 2005, 2010; Gaydos and Miller, 2008; Lasek-Nesselquist et al., 2008, 2010). Our findings indicate that assemblage B is the most common assemblage detected in wild and captive sea lions while assemblage A occurs at low frequency. The host range of assemblages A and B are broad including domestic animals, livestock and humans (reviewed in Monis et al., 1999; Monis and Thompson, 2003; Monis et al., 2003). Infections with assemblages A and B are very common in human cases but based on the absence of subtype characterization, it is difficult to assess the association between parasite transmission and humans.
The presence of G. duodenalis assemblage AI and B in this sea lion species is a strong indicator of the spread of parasites from terrestrial mammals to the marine environment. Giardia duodenalis is reported in higher frequencies in seal and sea lion species distributed within close proximity to human settlements and wastewater runoff localities (Gaydos and Miller, 2008; Appelbee et al., 2010). The presence of G. duodenalis in seals visiting haul-out sites distributed near coastal settlements can be up to five times greater than individuals at more sparsely populated sites with limited human exposure (Hughes-Hanks et al., 2005; Dixon et al., 2008; Gaydos and Miller, 2008; Lasek-Nesselquist et al., 2010). Some Australian sea lion colonies are within close proximity to coastal settlements and experience high levels of human interaction as a major tourist icon (Gales et al., 1994; Rodger et al., 2011). Sea lion behaviour such as hauling-out on human impacted beaches increases the potential for exposure to parasites from terrestrial sources. Compared with the more isolated Australian sea lion colonies (>70 km from human settlement), Giardia presence is significantly higher in colonies nearer (<25 km) to human coastal settlements and those colonies that experience high human visitation. Australian sea lions have limited dispersal and a high degree of philopatry so impacts are likely to be localized (Lowther et al., 2012). Future observation of protozoan prevalence in South Australian (Seal Bay and Seal Slide on Kangaroo Island) and Western Australian colonies (Shoalwater Marine Park, Perth; North Fisherman Island, Jurien Bay and Recherche Archipelago, Esperance) is therefore essential for monitoring the spread of parasites and associated potential disease risks, and will assist in the development of conservation management strategies.
Giardia duodenalis presence is significantly greater in captive Australian sea lions (36.8%) than wild animals (10.3%) indicating that occurrence may be the result of atypical habitat interactions. Exposure to humans and interactions atypical to those within the natural habitat of sea lions may increase the risk of Giardia transmission within captive environments (Beck et al., 2011). Captive mammals may be exposed to Giardia through human contact during hand feeding and touching by zoo visitors (Thompson et al., 2008). The captive facilities observed in this study have varying levels of visitor interaction programs with sea lions, some even include activities such as swimming with and touching the animals. While Giardia presence in captive marine mammal populations is rarely observed or indeed investigated, screening of Giardia in other captive mammal species with similar levels of human interactions would provide an indication of the extent of transmission in the captive environment. This in turn may reveal potential avenues of dispersal of Giardia in the captive environment, and by deduction, potential mitigation strategies for improved husbandry.
While the use of molecular tools has facilitated a greater understanding of protozoan origins and host specificity, we had limited success in accurately confirming Giardia species sub-assemblage across multiple loci. This poses significant biological implications for inferring host specificity and transmission of Giardia. We failed to amplify 18S rRNA positive isolates at the gdh and β-giardin loci. Difficulty in confirming positive 18S rRNA detection at the gdh locus has been observed in other marine and captive mammal studies. Failure to amplify at the gdh locus in samples from Pacific harbor seals and captive mammals was attributed to variation in sequences and failure of primers to anneal (Lasek-Nesselquist et al., 2010; Beck et al., 2011). While analyses at the 18S rRNA locus alone can enable assemblage identification, multilocus gene screening is required to determine G. duodenalis sub-assemblage and specific host origin.
Further, we were unable to consistently assign G. duodenalis assemblage across multiple loci for the one sample that amplified at β-giardin. Inconsistent assemblage identification across multiple loci has been observed in other marine mammal studies (Lasek-Nesselquist et al., 2008, 2010). Failure to confirm genotype across multiple loci in samples from grey and Pacific harbor seals was attributed to target gene amplification biases, where assemblages A and B preferentially amplified at different loci, and the presence of mixed assemblage infection (Lasek-Nesselquist et al., 2010). Mixed infection of G. duodenalis assemblages A and B are commonly reported in human and marine mammal studies, although there is much debate about whether this is the result of infection by multiple isolates or the haplotype of a single isolate (Cacciò and Ryan, 2008; Cacciò et al., 2008; Lasek-Nesselquist et al., 2008, 2010; Feng and Xiao, 2011). While the occurrence of recombination between G. duodenalis assemblages A and B has been supported by several studies, the mechanisms involved remain unclear (Cooper et al., 2007; Teodorovic et al., 2007; Cacciò and Ryan, 2008; Cacciò et al., 2008; Cacciò and Sprong, 2010; Lasek-Nesselquist et al., 2010). Consequently, due to limited amplification across multiple loci, we were unable to draw inferences on the potential for target gene amplification biases or the presence of mixed infection. These findings emphasize the need for multilocus molecular characterization to definitively assign G. duodenalis assemblages present in wild sea lion populations and determine the origin of parasite dispersal into the marine environment.
Increasing exposure to agricultural run-off and untreated wastewater represents new challenges for managing the dispersal of protozoan parasites into the marine ecosystem. The high occurrence of Giardia and similarity to Giardia species found in humans in both wild and captive sea lions warrants the need for further molecular investigation to identify the dispersal routes of parasites from terrestrial ecosystems into marine vertebrate populations.
Conflict of interest
The authors declared that there is no conflict of interest. All authors and relevant institutions have been acknowledged for their contributions.
Acknowledgements
This research was conducted under permission of the Department for Environment and Heritage, South Australia (permit Z25675) and DEH SA Wildlife Ethics Committee (44/2008). We thank the Department of Environment and Conservation, Western Australia (DEC WA License SF007255 and SF008193) for permission to enter the study sites in Western Australia. The research was supported by grants to Robert Harcourt from the Australian Marine Mammal Centre, Department of Environment, Heritage South Australia and West Australia, Water and the Arts through the Commonwealth Environment Research Fund and Macquarie University (0809/27, 09/38, 0809/12). Funding was also received from Macquarie University Department of Biological Sciences postgraduate project support to Tiffany Delport and Michelle Power. We would like to thank Dolphin Marine Magic, Sea World Gold Coast and Taronga Zoo for kindly providing Australian sea lion faecal samples and also Ben Pitcher, Andy Lowther, Isabelle Charrier and Heidi Ahonen for their help with wild animal faecal collection. We would also like to thank Prof. John Ellis and Dr. Sonja Frolich from the University of Technology, Sydney, for kindly providing Giardia trophozoite cultures used in this study.
References
- Appelbee A.J., Thompson R.C.A., Olson M.E. Giardia and Cryptosporidium in mammalian wildlife – current status and future needs. Trends Parasitol. 2005;21:370–376. doi: 10.1016/j.pt.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbee A.J., Thompson R.C.A., Measures L.M., Olson M.E. Giardia and Cryptosporidium in harp and hooded seals from the Gulf of St. Lawrence, Canada. Vet. Parasitol. 2010;173:19–23. doi: 10.1016/j.vetpar.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Asher A.J., Waldron L.S., Power M.L. Evaluation of a PCR protocol for sensitive detection of Giardia intestinalis in human faeces. Parasitol. Res. 2012;110:853–858. doi: 10.1007/s00436-011-2565-3. [DOI] [PubMed] [Google Scholar]
- Bass A.L., Wallace C.C., Yund P.O., Ford T.E. Detection of Cryptosporidium sp. in two new seal species, Phoca vitulina and Cystophora cristata, and a novel Cryptosporidium genotype in a third seal species, Pagophilus groenlandicus, from the Gulf of Maine. J. Parasitol. 2012;98:316–322. doi: 10.1645/GE-2853.1. [DOI] [PubMed] [Google Scholar]
- Beck R., Sprong H., Bata I., Lucinger S., Pozio E., Cacciò S.M. Prevalence and molecular typing of Giardia spp. in captive mammals at the zoo of Zagreb, Croatia. Vet. Parasitol. 2011;175:40–46. doi: 10.1016/j.vetpar.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Bogomolni A.L., Gast R.J., Ellis J.C., Dennett M.R., Pugliares K.R., Lentell B.J. Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis. Aquat. Organ. 2008;81:13–38. doi: 10.3354/dao01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciò S.M., Ryan U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Cacciò S.M., Sprong H. Giardia duodenalis: genetic recombination and its implications for taxonomy and molecular epidemiology. Exp. Parasitol. 2010;124:107–112. doi: 10.1016/j.exppara.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Cacciò S.M., De Giacomo M., Pozio E. Sequence analysis of the β-giardin gene and development of a polymerase chain reaction–restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 2002;32:1023–1030. doi: 10.1016/s0020-7519(02)00068-1. [DOI] [PubMed] [Google Scholar]
- Cacciò S.M., Thompson R.C., McLauchlin J., Smith H.V. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Cacciò S.M., Beck R., Lalle M., Marinculic A., Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Adam R.D., Worobey M., Sterling C.R. Population genetics provides evidence for recombination in Giardia. Curr. Biol. 2007;17:1984–1988. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Deng M., Peterson R.P., Cliver D.O. First findings of Cryptosporidium and Giardia in California sea lions (Zalophus californianus) J. Parasitol. 2000;86:490–494. doi: 10.1645/0022-3395(2000)086[0490:FFOCAG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dixon B.R., Parrington L.J., Parenteau M., Leclair D., Santin M., Fayer R. Giardia duodenalis and Cryptosporidium Spp. in the intestinal contents of ringed seals (Phoca hispida) and bearded seals (Erignathus barbatus) in Nunavik, Quebec, Canada. J. Parasitol. 2008;94:1161–1163. doi: 10.1645/GE-1485.1. [DOI] [PubMed] [Google Scholar]
- ESRI Inc . 2010. ArcGIS. Version 10.0. Redlands, CA. [Google Scholar]
- Feng Y., Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and Giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gales N.J., Shaughnessy P.D., Dennis T. Distribution, abundance and breeding cycle of the Australian sea-lion Neophoca cinerea (Mammalia: Pinnipedia) J. Zool. 1994;234:353–370. [Google Scholar]
- Gaydos J.K., Miller W.A. Novel and canine genotypes of Giardia duodenalis in harbor seals (Phoca vitulina richardsi) J. Parasitol. 2008;94:1264–1268. doi: 10.1645/GE-1321.1. [DOI] [PubMed] [Google Scholar]
- Goldsworthy S., Gales N. IUCN 2011. IUCN Red List of Threatened Species. Version 2011.1. 2008. Neophoca cinerea.http://www.iucnredlist.org accessed 08.06.30. [Google Scholar]
- Goldsworthy S.D., Shaughnessy P.D., Page B., Dennis T.E., MacIntosh R.R., Hamer D. Developing population monitoring protocols of Australia sea-lions. Final report to the Department of the Environment and Water Resources. SARDI Res. Rep. Ser. 2007;219:75. [Google Scholar]
- Hopkins R.M., Meloni B.P., Groth D.M., Wetherall J.A., Reynoldson R.C., Thompson A. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- Hueffer K., Holcomb D., Ballweber L.R., Gende S.M., Blundell G., O'Hara T.M. Serologic surveillance of pathogens in a declining harbor seal (Phoca vitulina) population in Glacier Bay National Park, Alaska, USA and a reference site. J. Wildl. Dis. 2011;47:984–988. doi: 10.7589/0090-3558-47.4.984. [DOI] [PubMed] [Google Scholar]
- Hughes-Hanks J.M., Rickard L.G., Panuska C., Sauciert J.R., O'Harat T.M., Dehn L. Prevalence of Cryptosporidium spp. and Giardia spp. in five marine mammal species. J. Parasitol. 2005;91:1225–1228. doi: 10.1645/GE-545R.1. [DOI] [PubMed] [Google Scholar]
- Kosuwin R., Putaporntip C., Pattanawong U., Jongwutiwes S. Clonal diversity in Giardia duodenalis isolates from Thailand: evidences for intragenic recombination and purifying selection at the beta giardin locus. Gene. 2010;449:1–8. doi: 10.1016/j.gene.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Lalle M., Pozio E., Capelli G., Bruschi F., Crotti D., Cacciò S.M. Genetic heterogeneity at the β-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., Bogomolni A.L., Gast R.J., Welch D.M., Ellis J.C., Sogin M.L. Molecular characterization of Giardia intestinalis haplotypes in marine animals: variation and zoonotic potential. Dis. Aquat. Organ. 2008;81:39–51. doi: 10.3354/dao01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., Welch D.M., Sogin M.L. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. J. Parasitol. 2010;40:1063–1074. doi: 10.1016/j.ijpara.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbad M., Mattsson J.G., Christensson B., Ljungström B., Backhans A., Andersson J.O. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet. Parasitol. 2010;168:231–239. doi: 10.1016/j.vetpar.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Lowther A.D., Harcourt R.G., Goldsworthy S.D., Stow A. Population structure of adult female Australian sea lions is driven by fine-scale foraging site fidelity. Anim. Behav. 2012;83:691–701. [Google Scholar]
- Monis P.T., Thompson R.C. Cryptosporidium and Giardia zoonoses: fact or fiction? Infect. Genet. Evol. 2003;3:233–244. doi: 10.1016/j.meegid.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Monis P.T., Andrews R.H., Mayrhorfer G., Ey P.L. Molecular systematics of the parasitic protozoan Giardia intestinalis. Mol. Biol. Evol. 1999;16:1135–1144. doi: 10.1093/oxfordjournals.molbev.a026204. [DOI] [PubMed] [Google Scholar]
- Monis P.T., Andrews R.H., Mayrhorfer G., Ey P.L. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect. Genet. Evol. 2003;3:29–38. doi: 10.1016/s1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
- Olson M.E., Roach P.D., Stabler M., Chan W. Giardiasis in ringed seals from the Western Arctic. J. Wildl. Dis. 1997;33:646–648. doi: 10.7589/0090-3558-33.3.646. [DOI] [PubMed] [Google Scholar]
- Read C.M., Monis P.T., Andrew Thompson R.C. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect. Genet. Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Rengifo-Herrera C., Ortega-Mora L.M., Gómez-Bautista M., García-Moreno F.T., García-Párraga D., Castro-Urda J. Detection and characterization of a Cryptosporidium isolate from a Southern elephant seal (Mirounga leonina) from the Antarctic Peninsula. Appl. Environ. Microbiol. 2011;77:1524–1527. doi: 10.1128/AEM.01422-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengifo-Herrera C., Ortega-Mora L.M., Gómez-Bautista M., García-Peña F.J., García-Párraga D., Pedraza-Díaz S. Detection of a novel genotype of Cryptosporidium in Antarctic pinnipeds. Vet. Parasitol. 2013;191:112–118. doi: 10.1016/j.vetpar.2012.08.021. [DOI] [PubMed] [Google Scholar]
- Rodger K., Smith A., Newsome D., Moore S.A. Developing and testing an assessment framework to guide the sustainability of the marine wildlife tourism industry. J Ecotourism. 2011;10:149–164. [Google Scholar]
- Ryan U., Fayer R., Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014:1–19. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Santin M., Dixon B.R., Fayeri R. Genetic characterization of Cryptosporidium isolates from ringed seals (Phoca hispida) in Northern Quebec, Canada. J. Parasitol. 2005;91:712–716. doi: 10.1645/GE-3438RN. [DOI] [PubMed] [Google Scholar]
- Shaughnessy P.D., Goldsworthy S.D., Hamer D., Page B., McIntosh R.R. Australian sea lions Neophoca cinerea at colonies in South Australia: distribution and abundance, 2004 to 2008. Endanger Species Res. 2011;13:87–98. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodorovic S., Braverman J.M., Elmendorf H.G. Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryot. Cell. 2007;6:1421–1430. doi: 10.1128/EC.00138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Yang R., Power M., Hufschmid J., Beveridge I., Reid S. Identification of zoonotic Giardia genotypes in marsupials in Australia. Exp. Parasitol. 2008;120:88–93. doi: 10.1016/j.exppara.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, population-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron L.S., Dimeski B., Beggs P.J., Ferrari B.C., Power M.L. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human Cryptospordiosis in Australia. Appl. Environ. Microbiol. 2011;77:7757–7765. doi: 10.1128/AEM.00615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielinga C.M., Thompson R.C.A. Comparative evaluation of Giardia duodenalis data. Parasitology. 2007;134:1795–1821. doi: 10.1017/S0031182007003071. [DOI] [PubMed] [Google Scholar]
- Xiao L. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Alderisio K., Limor J., Royer M., Lal A.A. Identification of species and sources of Cryptosporidium oocysts in Storm Waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 2000;66:5492–5498. doi: 10.1128/aem.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


