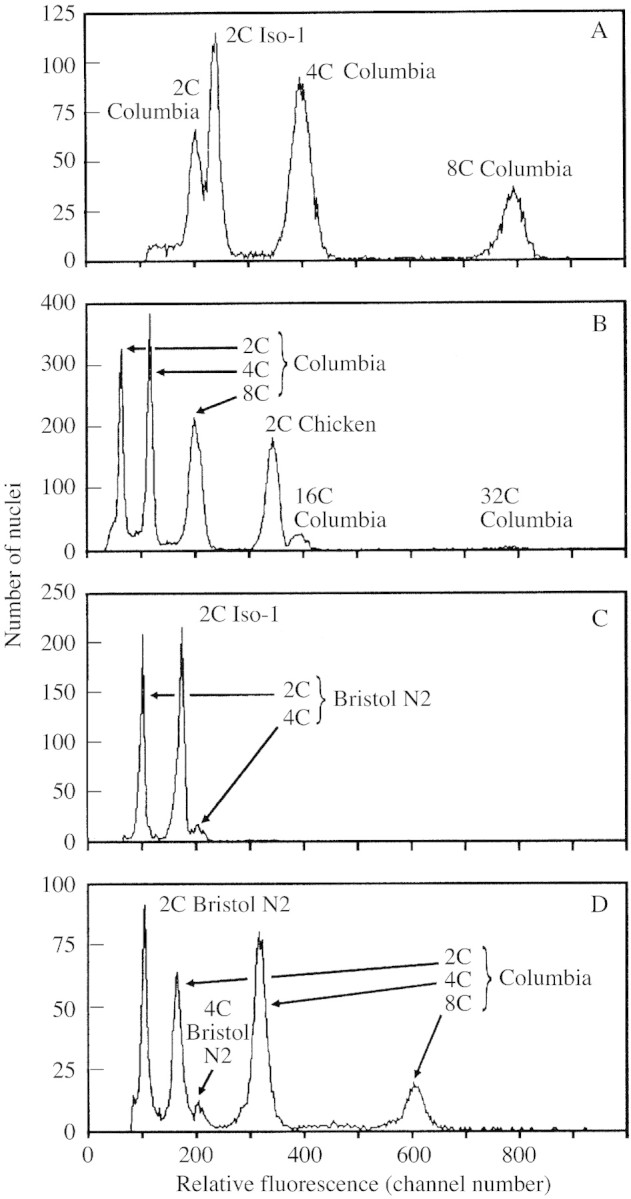
Fig. 1. Relative DNA staining in nuclei of arabidopsis, drosophila, Gallus and Caenorhabditis. A, Arabidopsis has approx. 85 % of the 2C nuclear DNA fluorescence of drosophila. Relative red fluorescence of PI‐stained nuclei shows a ratio of approx. 0·85 : 1·00 between A. thaliana ecotype Columbia (2C mean = 201·6) and female D. melanogaster strain Iso‐1 (2C mean = 236·7). The arabidopsis 2C peak was identified as the lowest in its 2C (mean = 201·6), 4C (mean = 398·8), 8C (mean = 792·1), etc. doubling series. PI staining time = 4 h. B, Arabidopsis 16C nuclear DNA fluorescence exceeds that of 2C chicken. Relative red fluorescence of PI‐stained nuclei shows a ratio of approx. 1·15 : 1·00 between endopolyploid 16C nuclei of A. thaliana ecotype Columbia (16C peak mean = 390·9) and chicken erythrocyte nuclei (2C peak = 341·7). NB Arabidopsis is identified from its known doubling series 2C, 4C, 8C, 16C.) PI staining time = 2·8 h. C, Drosophila has approx. 175 % 2C nuclear DNA fluorescence of C. elegans. Relative red fluorescence of PI‐stained nuclei shows a ratio of approx. 1·00 : 1·74 between C. elegans variety Bristol N2 (2C mean = 95·9) and female D. melanogaster strain Iso‐1 (2C mean = 167·2). PI staining time = 9·3 h. D, Arabidopsis has approx. 157 % 2C nuclear DNA fluorescence of Caenorhabditis. Relative red fluorescence of PI‐stained nuclei shows a ratio of approx. 1·00 : 1·54 between C. elegans variety Bristol N2 (2C mean = 101·6) and A. thaliana ecotype Columbia (2C mean = 159·6). PI staining time = 4·1 h. NB Species were identified by varying the relative amounts of nuclei from one to the other between runs within each mixture.
