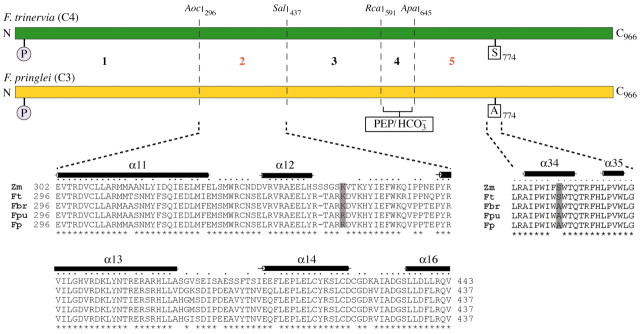
Fig. 5. An evolutionary model of C4 PEPC in Flaveria. From five investigated enzyme domains, region 2 (positions 296–437) and region 5 (amino acids 645–966) contain the major C4 determinants for the saturation kinetics of PEP. P indicates the target phosphorylation site at position 11. The secondary structures indicated on top of the sequence alignments (black bars) were obtained from the recently published 3D structure of the C4 PEPC of Zea mays (Matsumura et al., 2002). The dots above the maize sequence show those amino acid residues that are conserved in all displayed sequences. Sequence positions which are identical in all four Flaveria ppcA PEPCs are marked by stars below the strings of sequences. Note that position 347 (grey column) of the PEPC isoenzymes harbours a lysine in the C4 species F. trinervia and Z. mays, and in the C4‐like plant F. brownii, while the C3 species F. pringlei and the C3–C4 intermediate plant F. pubescens both instead hold an arginine at this site. At position 774 (grey column) serine occurs only in C4 PEPCs, while PEPCs from C3 and C3–C4 intermediate plants contain an alanine at this position. The amino acid numbering follows that of the F. trinervia protein.
