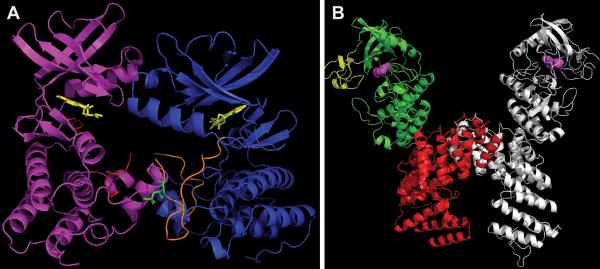
FIGURE 7.
Distinct modes of monomer interaction in dimers of PknB versus PknG (A) “Front-to-front” dimer of mutant PknB KD (3F69) in complex with Kt5720 inhibitor (yellow). The “substrate” subunit (magenta) has most of its activation loop disordered (red), while the “enzyme” subunit (blue) has a well-defined activation loop (orange) with visible phosphor-ylated Thr171 (green). (B) Structure of PknG (2PZI) in complex with inhibitor Ax20017 (magenta). Three domains: rubredoxin (yellow), KD (green), and TPR domain (red) are shown only in one subunit. The second subunit is depicted in gray. Figures were made in PyMOL (Schrödinger) and POV-Ray (povray.org). doi:10.1128/microbiolspec.MGM2-0006-2013.f7