Figure 1.
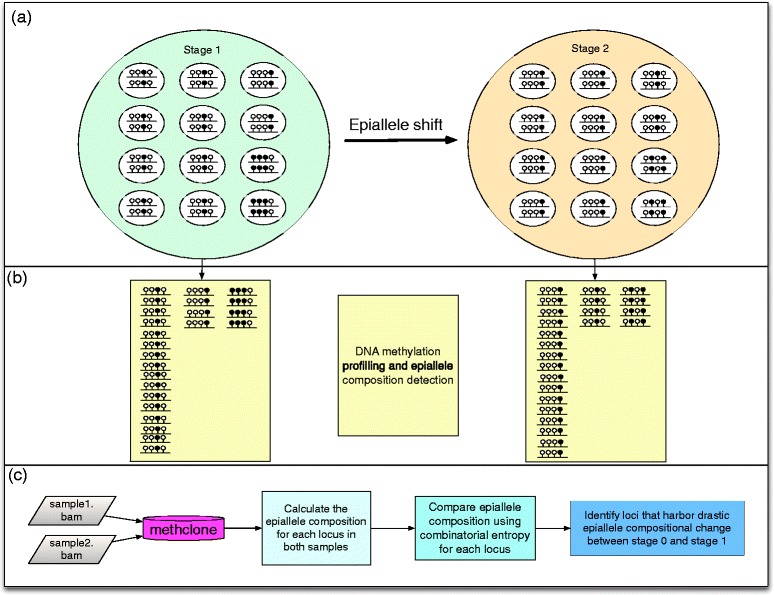
Epiallele shift detection by methclone. (a) Schematic plot of epiallele composition of two stages of cells (biggest circles, light green for stage 1 and light brown for stage 2). Each stage has 12 cells with various epiallele compositions. In each cell, there are two lines, represent two set of epiallele at the same locus. Four circles above each line represent four adjacent CpG sites (black: methylated CpG; white: unmethylated CpG). (b) Bisulfite conversion sequencing output reads that spanning at least four CpG sites will capture the epiallele composition at each stage. (c) methclone workflow. methclone take the bam file from Bismark to calculate the epiallele composition and compare them from different samples. In the application of the combinatorial entropy, methclone determine the loci that harbor significant epiallele compositional change.
