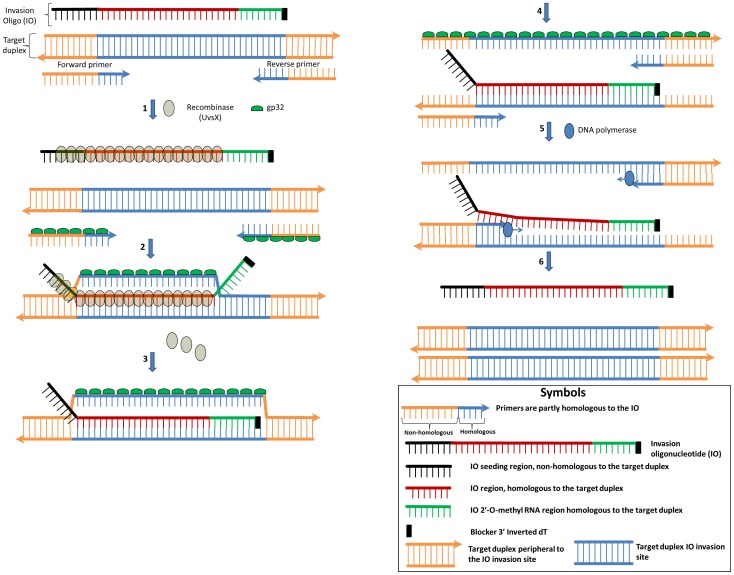
Figure 1. Mechanistic description of the SIBA reaction.
All single-stranded elements are coated with gp32, except for the 2′-O-methyl RNA nucleotides. Step 1: UvsX displaces gp32 on the IO and only weakly coats the primers, since they are too short for high affinity binding. Step 2: The IO invades the complementary region of the target duplex which allows partial separation of the target duplex with the downstream end still remaining double-stranded. The out-going strand of the partially separated target duplex is stabilized by gp32. Step 3: UvsX depolymerization allows the 2′-O-methyl RNA region of the IO branch migrates into the duplex. Step 4: Both the upstream and downstream region peripheral to the IO also become short enough to dissociate. Step 5: The strand displacement polymerase is able to extend the dissociated target duplex from the primers. The forward primer displaces the IO during extension of the target template. Step 8: These events lead to the production of two copies of the target duplex. The IO is released to induce further amplification.