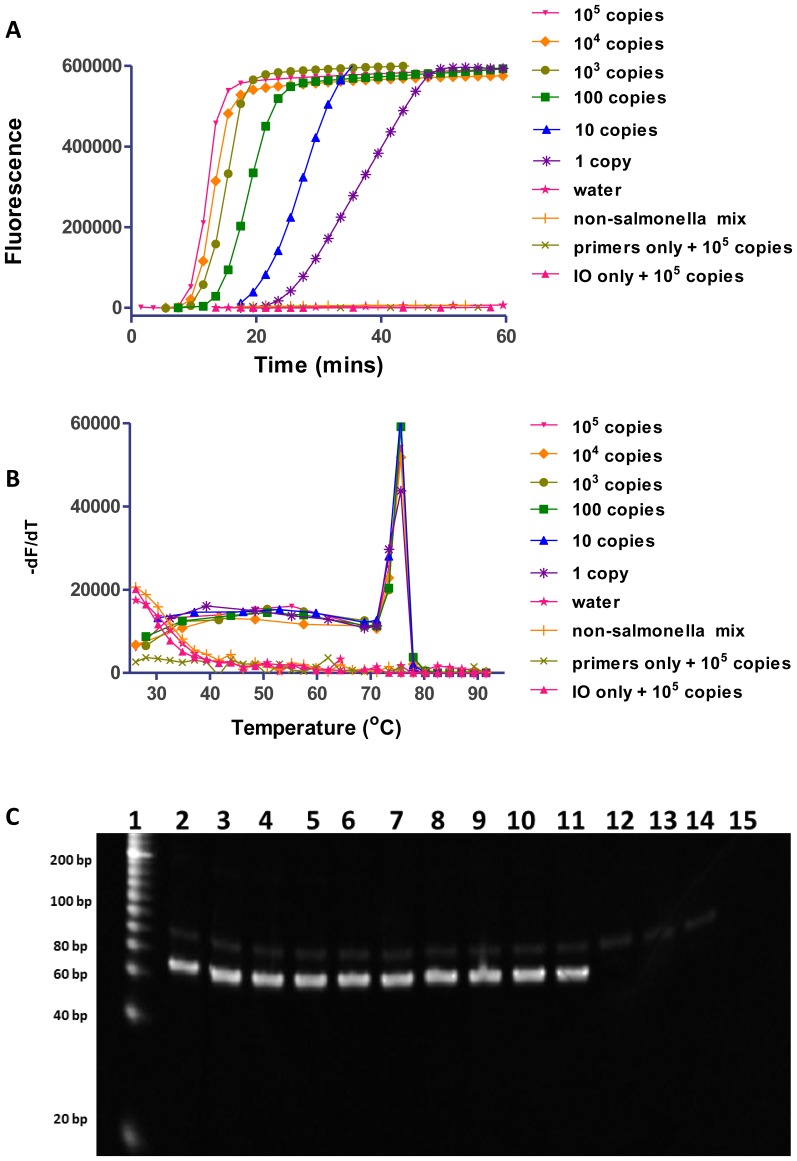
Figure 5. Determination of SIBA sensitivity for Salmonella genomic DNA.
(A) Real-time monitoring of DNA amplification using SYBR Green I, (B) melting curve analysis, ((-dF (fluorescence)/dT (temperature) versus temperature) and (C) electrophoresis of the corresponding reaction products. Lane 1, BioRad EZ Load 20 bp Molecular Ruler (20–1000 bp); lanes 2–11 primers + IO + 105, 104, 103, 100, 50, 20, 10, 5, 2, or 1 copy of Salmonella genomic DNA, respectively; lane 12, primers + IO + water; lane 13, primers + IO + a mixture of non-Salmonella spp. genomic DNA (each spp. contains 1000 copies per reaction); lane 14, IO + 105 copies Salmonella genomic DNA; lane 15, primers + 105 copies Salmonella genomic DNA; 50, 20, 5 and 2 copies were omitted from Figure 5a and b for the sake of clarity. SM-F18 and SM-R16 were the forward and reverse primers, respectively. The IO used was SM-IO.