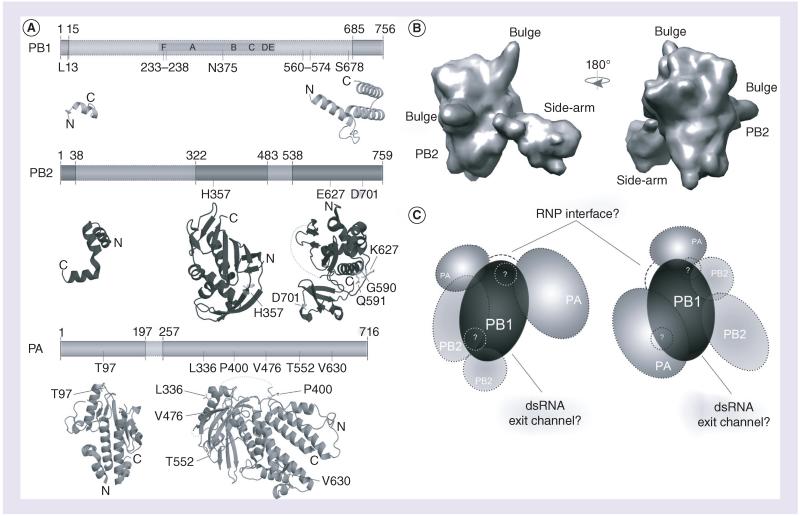
Figure 2. Structure of the influenza A virus RNA-dependent RNA polymerase.
(A) The influenza A virus (IAV) RNA-dependent RNA polymerase (RdRp) consists of the subunits PA, PB1 and PB2. Six of the seven canonical RdRp domains motifs A–F are found in PB1. Also indicated are the RNA binding sequences (gray), as are the residues involved in adaptation (black; see also Table 1). At present, significant structural information is only available for PA and PB2, which is shown below the schematics of the subunits. The visualized structural information is based on Protein Data Bank (PDB) entries 2VY6, 2W69, 2ZNL, 2ZTT, 3EBJ and 4CB4. (B) Two views of the electron microscopy model proposed by Moeller et al. [39]. Panels are based on the PDBe entry EMD-2213. (C) Two interpretations based on current electron microscopy models. On the left, the PA C-terminus presents the flexible part of the IAV RdRp, as defined by Moeller et al. [39]. It is connected to the PA endonuclease domain via a linker [37], which may wrap around PB1. An interaction through the core of PB1 would seem unlikely, as this may interfere with the right-handed fold of the polymerase active site. On the right, the PB2 C-terminus presents the flexible part of the IAV RdRp, as proposed by Torreira et al. [49], The PA domains flank PB1 on the other side, but the PA N-terminus can still interact with PB2 in accordance with recent data [47]. In both models, the flexible domain can be envisioned as blocking the dsRNA exit channel of the RdRp, which likely resides opposite – albeit at a slight angle – to the template entry grove. Both interpretations also have a similar interface consisting of PB1 and PA residues in order to interact with the RNP. Since the PA linker domain may not be part of this interaction, it is displayed in both models as a dashed line. The areas in (C) labeled with question marks refer to the bulge structures in (B), whose identity is current unknown.
PA: Polymerase acidic; PB1: Polymerase basic 1; PB2: Polymerase basic 2; RNP: Ribonucleoprotein.