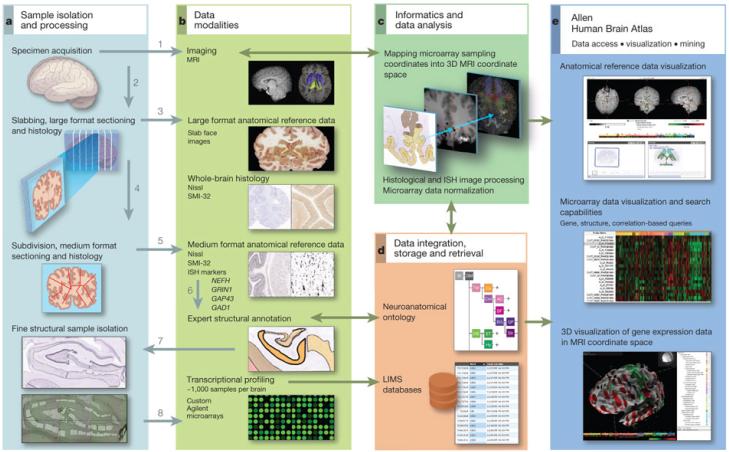
Figure 1. Data generation and analysis pipeline.
a, Experimental strategy to subdivide intact brains and isolate precise anatomical samples. b, Anatomical reference data are collected at each stage, including whole-brain MRI, large-format slab face and histology, medium (2 × 3-inch slide) format Nissl histology and ISH, and images of dissections. In Brain 2, labelling was performed for additional markers as shown. Histology data are used to identify structures, which are assembled into a database using a formal neuroanatomical ontology (d), and to guide laser microdissection of samples (a, lower panel). Isolated RNA is used for microarray profiling of ~900 samples per brain (b, lower panel). c, Microarray data are normalized and sample coordinates mapped to native 3D MRI coordinates. e, Data visualization and mining tools underlie the online public data resource. Numbers in a and b denote the order of sample processing steps leading to microarray data generation.