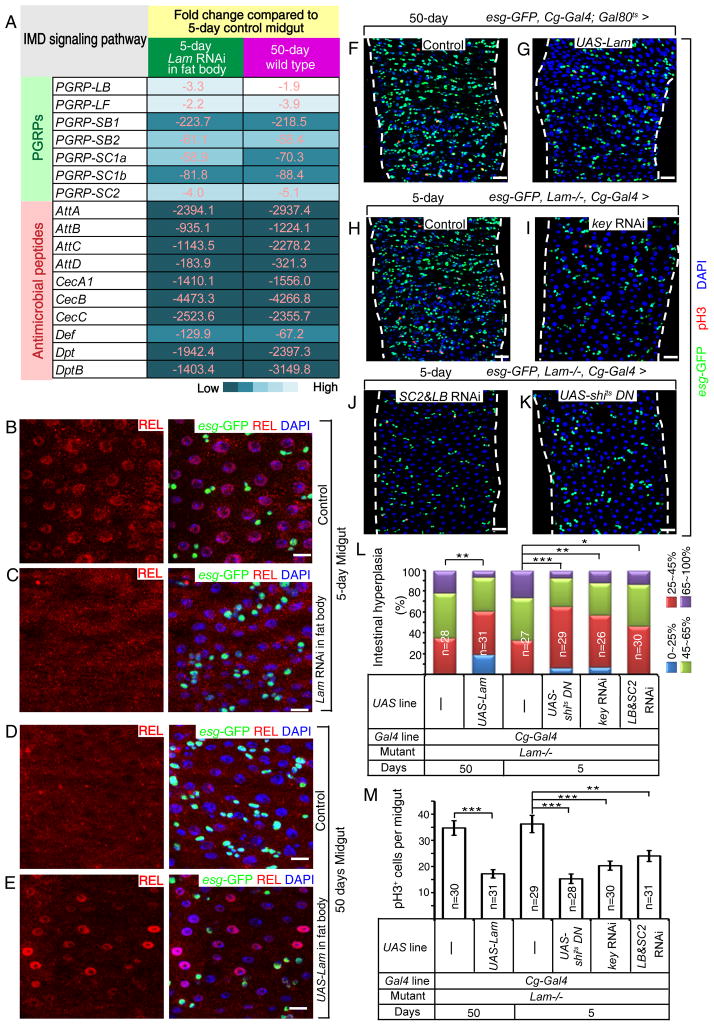
Figure 6. LAM maintains gut homeostasis by inhibiting systemic inflammation caused by the fat body.
A. A list of selected down-regulated IMD pathway genes in midguts from old wild-type flies or young flies raised in conventional condition with fat body-specific LAM depletion. Numbers and color shades indicate similar fold decrease.
B–E. In conventional condition, age-associated repression of IMD signaling in midgut (D) was reversed by fat body-specific expression of LAM in old flies (E) to levels similar to those of control young flies (B) or mimicked by fat body-specific Lam RNAi in young flies (C). REL, red; esg-GFP, green. DAPI (blue), nuclei. Scale bars, 10 μm.
F–K. Midgut hyperplasia in old flies (F) was inhibited by fat body-specific expression of LAM (G). Midgut hyperplasia in young Lam-mutant flies (H) was inhibited by fat body-specific RNAi of KEY (I), co-depletion of PGRP-SC2 & PGRP-LB (SC2 & LB, J), or fat body-specific expression of a dominant negative form of dynamin (shits) (K) in young Lam-mutant flies. Flies were raised in conventional condition. esg-GFP, green; pH3, red; DAPI, blue. Scale bars, 20 μm.
L. Midgut hyperplasia based on esg-GFP. The % of esg-GFP+ cells (% of intestinal hyperplasia) was grouped into four classes and plotted. *p<0.05, **p<0.01, ***p<0.001, Wilcoxon two-sample test.
M. Midgut cell proliferation based on pH3 staining. The average number of pH3+ cells was determined in whole midguts and plotted. Student’s t-tests, **p<0.01, ***p<0.001.
n, numbers of midguts analyzed. Error bars, SEM.