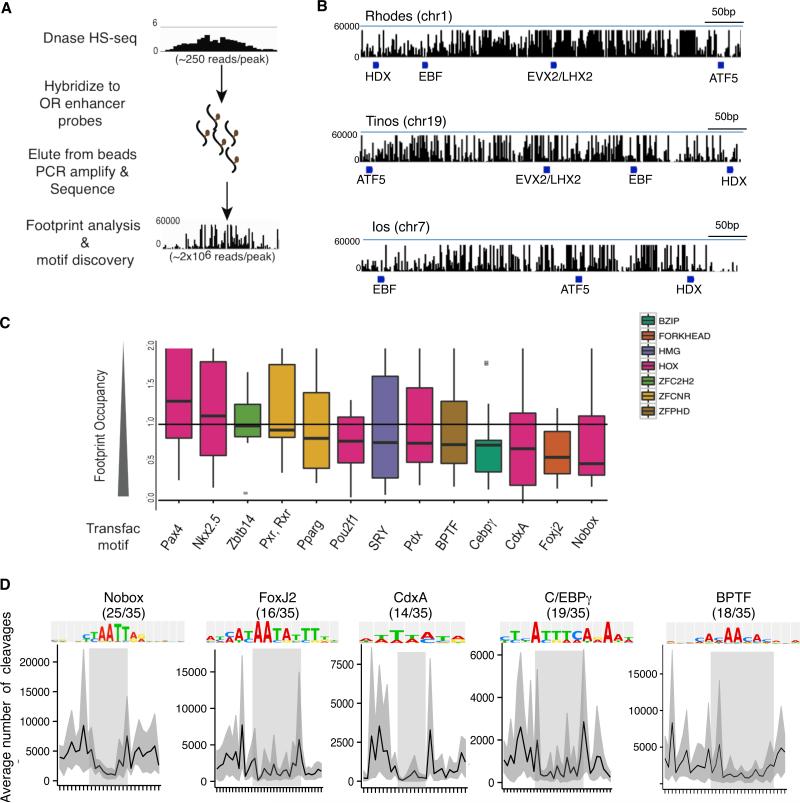
Figure 5. Shared transcription factor footprints on OR enhancers.
A. Schematic of sequence capture-based method of DNase I library construction.
B. DNase I cleavages mapped over OR enhancer candidates Rhodes, Tinos and Ios. Blue bars indicate footprints containing ATF5, HDX, EVX2/LHX2, and EBF motifs.
C. Transcription factor (TF) motifs from the TRANSFAC database that are enriched on OR enhancer candidates and average footprint score for each TF motif. Low FOS scores indicate greater footprint occupancy. TF motifs are color coded according to TF family.
D. Average DNase I cleavages for TRANSFAC motifs on 35 candidate OR enhancers. The X-axis is centered at the consensus sequence (shaded box). Fraction of OR enhancers containing each TF motif is indicated in parentheses. Error is bootstrapped 95% confidence intervals.
See also Figure S5 and Table S4 for footprint and motif locations.