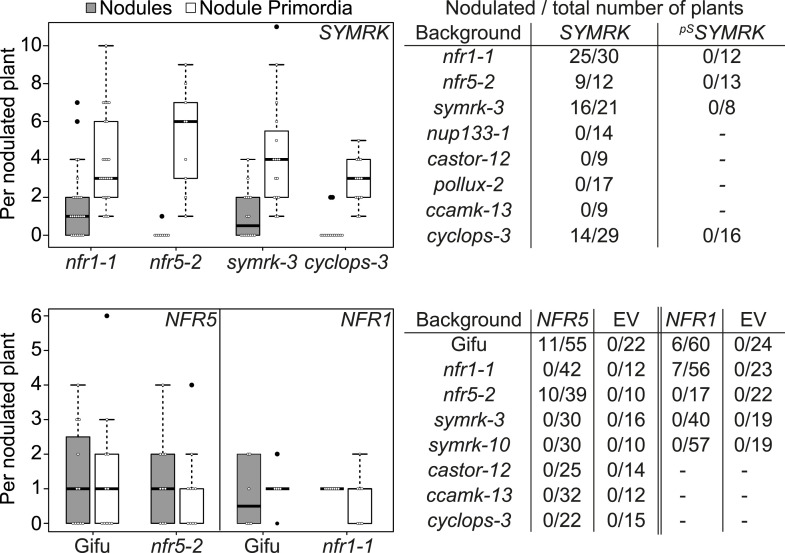
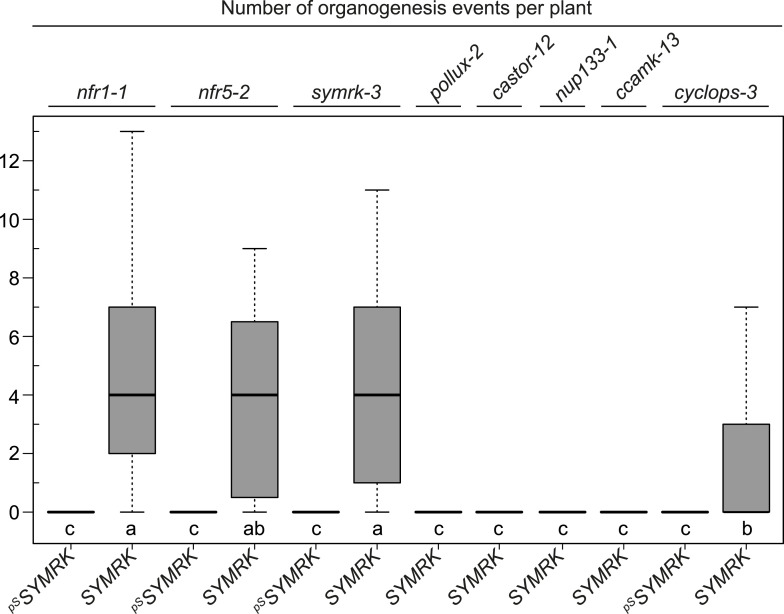
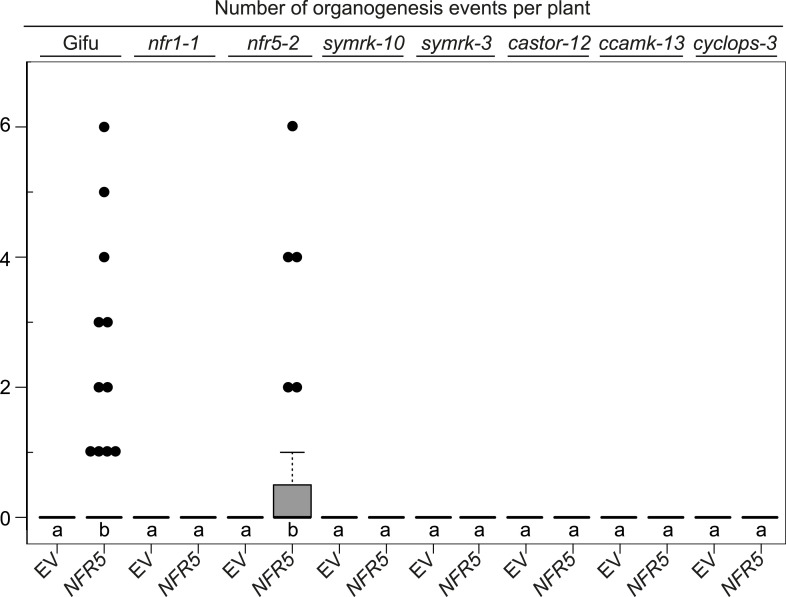
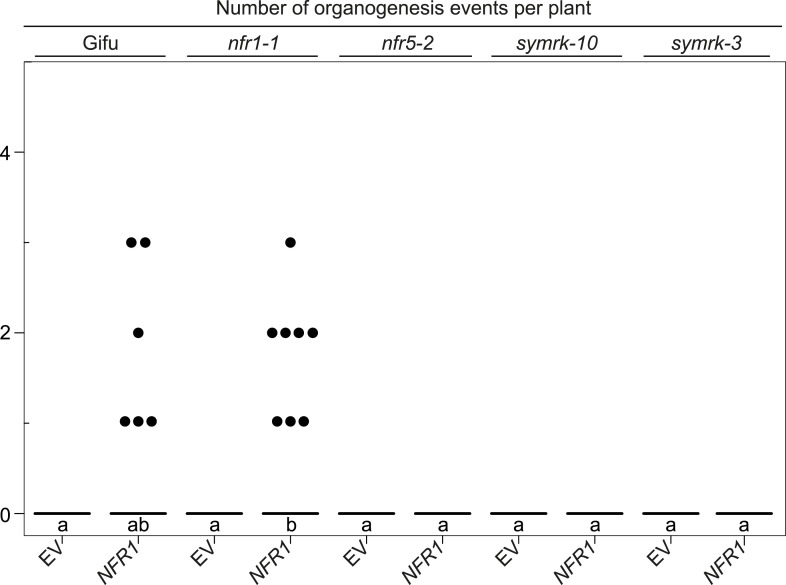
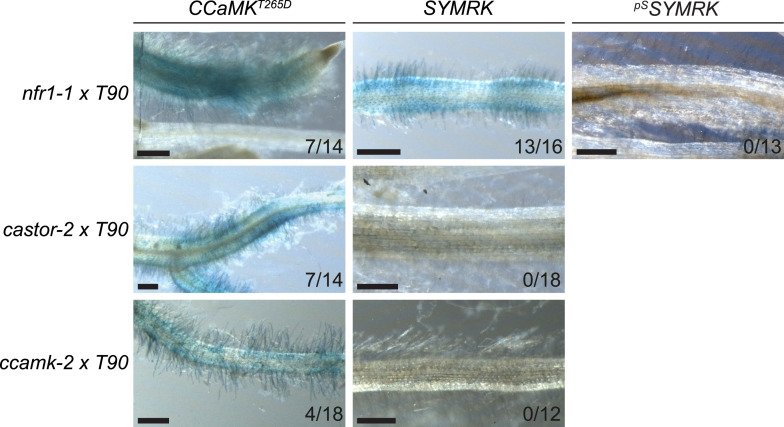
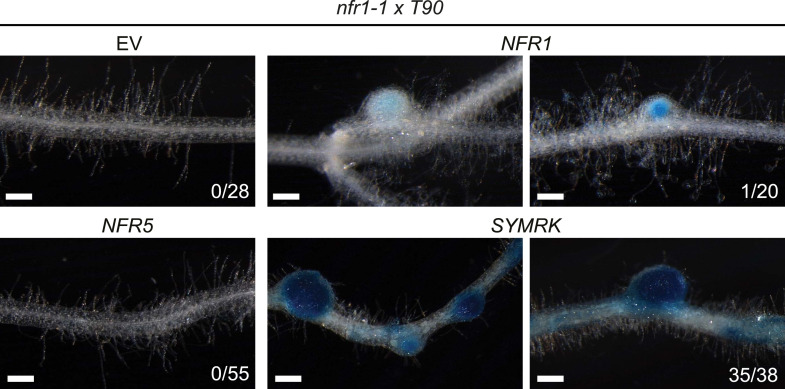
Figure 5. Epistatic relationships between symbiotic RLK genes and common symbiosis genes.
Hairy roots of L. japonicus Gifu wild-type and different symbiosis defective mutants transformed with pUB:SYMRK-mOrange (SYMRK) or pSYMRK:SYMRK-RFP (pS SYMRK) (upper panel), or the empty vector (EV), pUB:NFR1-mOrange (NFR1) or pUB:NFR5-mOrange (NFR5) (lower panel) were generated. Plots represent the numbers of nodules (grey) and nodule primordia (white) per nodulated plant formed in the absence of rhizobia at 40 (SYMRK) and 60 (NFR5 + NFR1) dpt. White circles indicate individual organogenesis events. Black dots, data points outside 1.5 IQR of the upper/lower quartile; bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. Table, fraction of nodulated per total number of plants. Plants transformed with pSYMRK:SYMRK-RFP or the empty pUB vector did not develop spontaneous nodules.