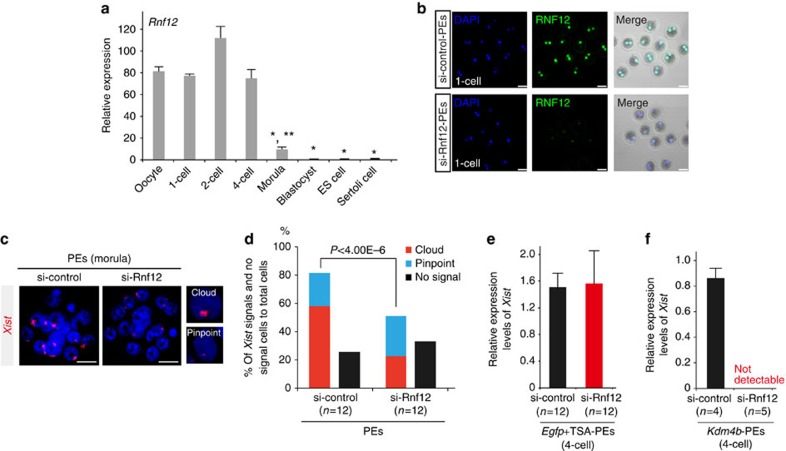
Figure 3. RNF12 is required for Xist expression in various types of preimplantation embryos.
(a) Rnf12 expression profiles in preimplantation stages, ES cells and somatic cells. Ten oocytes (n=3), ten IVF-1 cells (n=3), ten IVF-2 cells (n=3) and five IVF-4 cells (n=3) were pooled. The expression level of Rnf12 in the morula stage, as determined using qPCR, represents the average of 16 individual embryos. The numbers of ES and Sertoli cells represent the averages of three independent cell lines and male pups, respectively. The error bars indicate the mean±s.e.m. *P<0.004 compared with oocytes; **P<0.0001 compared with ES cells (Student’s t-test). (b) IF analysis of RNF12 at the one-cell stage (green). Samples were fixed 11 h after parthenogenetic activation (18–19 h after siRNA injection). The same laser beam intensities were used to excite the Rnf12-knockdown and control samples. 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei are shown in blue. Two independent experiments were conducted. Scale bars, 50 μm. (c,d) Xist FISH analysis of si-Rnf12 PEs at the morula stage. At 72 h after activation, PEs injected with siRNA were analysed. Representative images of siRNA-treated embryos (c). Scale bars, 20 μm. The percentage of total Xist-positive signals and -negative cells to total cells in si-Rnf12 and si-control PEs. Biallelic expression was counted as two signals. n, number of embryos analysed (d). (e,f) qPCR analysis of Xm-Xist expression at the four-cell stage of embryos treated with TSA (e) or injected with Kdm4b mRNA (f). PEs derived from maternal si-Rnf12-treated oocytes. A detailed experimental scheme is shown in Supplementary Fig. 6d. A pool of eight to ten four-cell embryos represents one biological replicate.