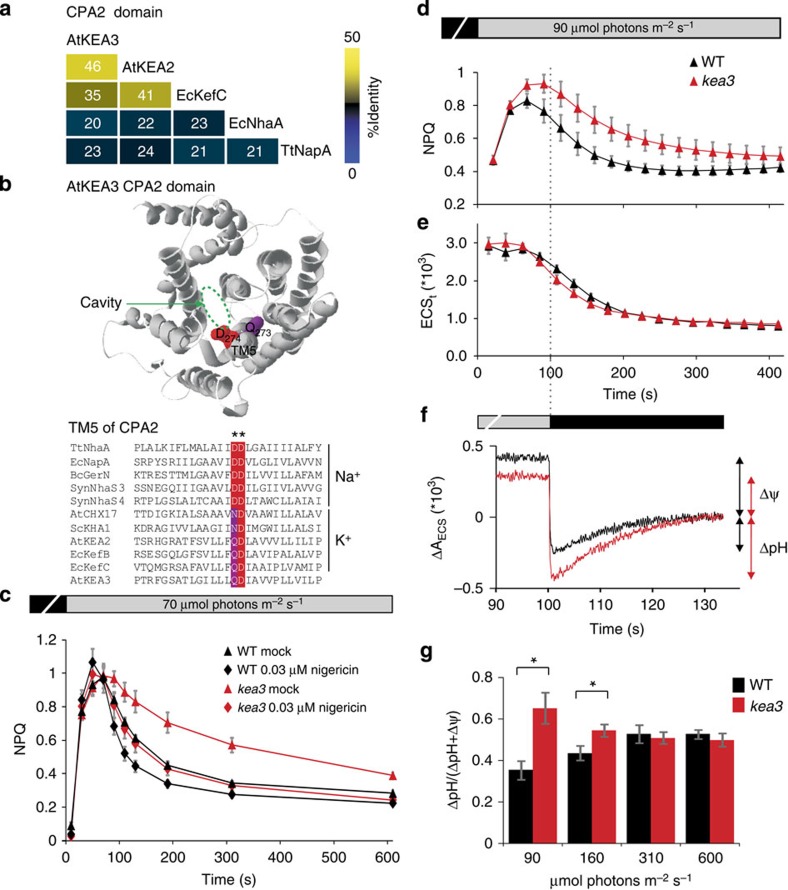
Figure 3. KEA3 regulates the composition of the p.m.f. by mediating potassium/proton antiport.
(a) The CPA2 domain of KEA3 is homologous to that of known K+/H+ antiporters. A percent identity matrix is shown for Arabidopsis KEA2 and KEA3, E. coli KefC and NhaA and T. thermophilus NapA. (b) The locations of two key substrate-binding amino acid residues in TM5 of the CPA2 domain19 are shown on a model of KEA3. (c) Low levels of Nigericin complement the kea3 NPQ phenotype. NPQ induction on transition from dark to 70 μmol m−2 s−1 light was measured in WT (Ws) and kea3-2 leaves incubated in water (mock) or 0.03 μM nigericin. (d,e) p.m.f. kinetics in kea3 mutants are largely unaffected. NPQ and ECSt (which reports the magnitude of the p.m.f.) were measured near-simultaneously in WT (Ws) and kea3-2 by Chl fluorescence and dark-induced relaxation kinetics, respectively, in single leaves during a transition from dark to 90 μmol m−2 s−1. (f) After 100 s of low light, kea3 mutants show increased ΔpH and decreased Δψ. Full ECS decay kinetics were recorded after 100 s of low light (90 μmol m−2 s−1) to measure ΔpH and Δψ in WT (Col-0) and kea3-1. The average of six independent measurements per genotype was plotted as a moving average with interval 5 (see also Supplementary Fig. 9). (g) The experiment in f was repeated at different light intensities, and the fraction of the p.m.f. contributed by ΔpH was plotted. Asterisks indicate where WT and kea3 differ significantly (*P<0.04, Student’s t-test). (c–e,g) Error bars represent s.e.m. (n=6).