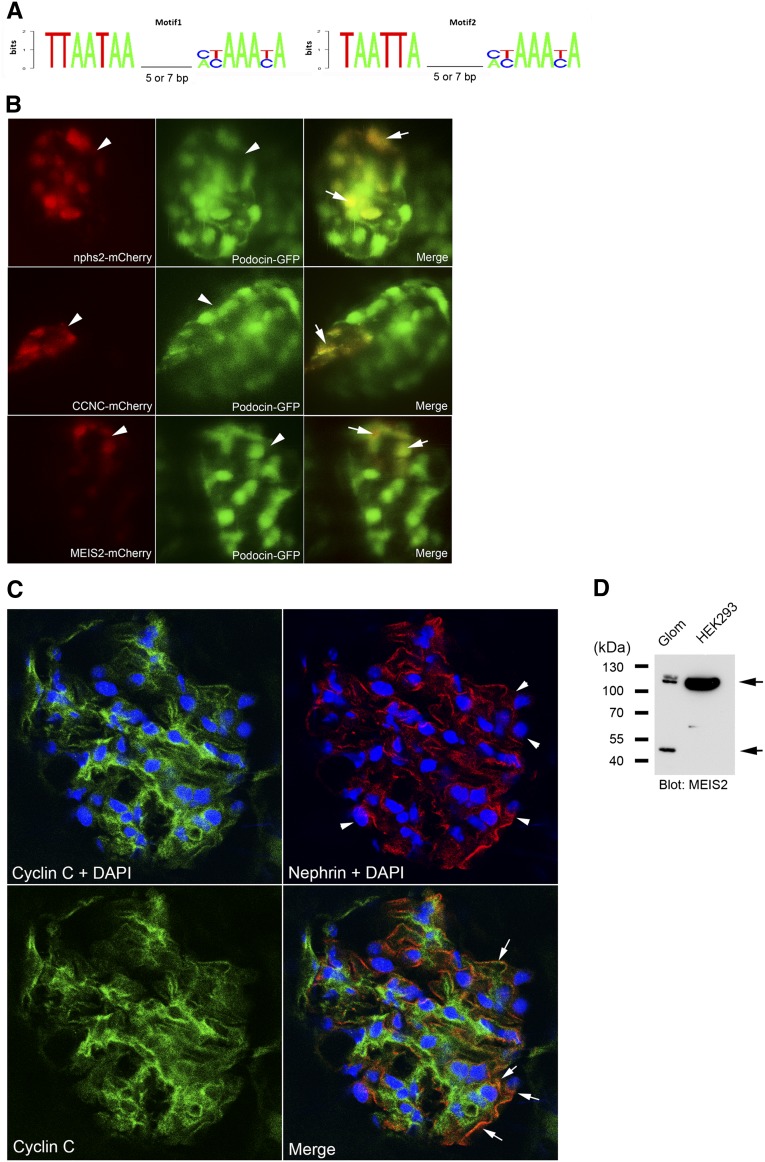
Figure 7.
Podocyte enhancer activity analyses of the predicted Lmx1b-FoxC motifs in CCNC and MEIS2 promoter. (A) Two sequence logos representing the position weight matrix of the Lmx1b-FoxC motif were used for the genome-wide search. Spacing sizes of 5 or 7 nucleotides between two sites were included. (B) In vivo assay for podocyte enhancer activity. Colocalization between transient mCherry expression and stable glomerular GFP expression in living zebrafish was analyzed using confocal microscopy. The zebrafish nphs2 enhancer was used as a positive control (upper panel). Mosaic mCherry expression (red, arrowhead) driven by the motifs of CCNC (middle panel) and MEIS2 (lower panel) shows colocalization (arrow) with glomerular GFP (green, arrowhead). (C) Confocal images of the mouse kidney section stained for cyclin C (green), nephrin (red), and nucleus with DAPI (blue). An image of cyclin C staining without DAPI (lower left panel) is included for assessment of nuclear staining in comparison to cyclin C staining with DAPI. Arrowhead indicates podocytes, and arrow indicates the overlap between cyclin C and nephrin. (D) In Western blotting, Meis2 was detected as two distinct bands (arrows), one with approximately 52-kD (predicted molecular mass, 52 kD) and one with about 110 kD, in mouse whole glomerular lysates (Glom). In nuclear extract of HEK293 cells, only a single strong band with about 110 kD was detected.