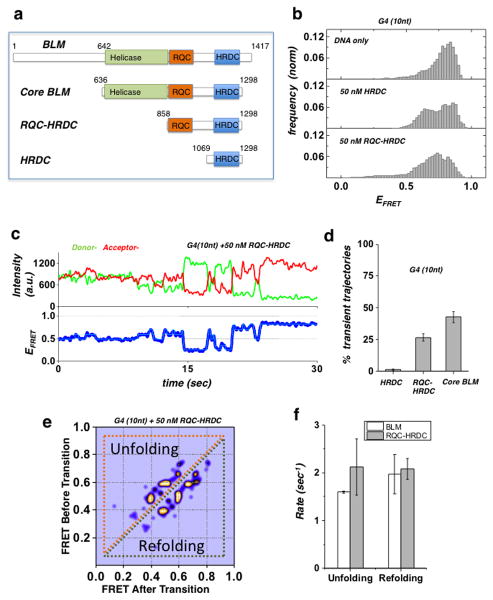
Figure 2. Domain analysis of BLM G4 unfolding activity.
(a) BLM protein functional domains and truncation fragments used in this study: full-length BLM, core BLM, RQC-HRDC fragment and HRDC.
(b) FRET histogram of the G4 (10nt) substrate showing DNA only substrate (top panel), and in the presence of 50 nM HRDC fragment (mid panel) and RQC-HRDC fragment (bottom panel).
(c) A representative smFRET trajectory of the G4 (10nt) substrate in the presence of 50 nM RQC-HRDC fragment, showing rapid FRET fluctuations (blue curve). HMM fit is in cyan.
(d) Quantification of population percentage of transient trajectories for the G4 (10 nt) substrate in the presence of 50 nM of HRDC and RQC-HRDC fragments and core BLM. (Error bar = S.E.M. n=4)
(e) TDP for the G4 (10 nt) substrate in the presence of 50 nM RQC-HRDC fragment.
(f) Quantification of the mean unfolding and refolding rates for the G4 motif of the G4 (10 nt) substrate, in the presence of 50 nM RQC-HRDC fragment or core BLM. (Error bar = S.E.M. n > 24 for all measurements)