Figure 5.
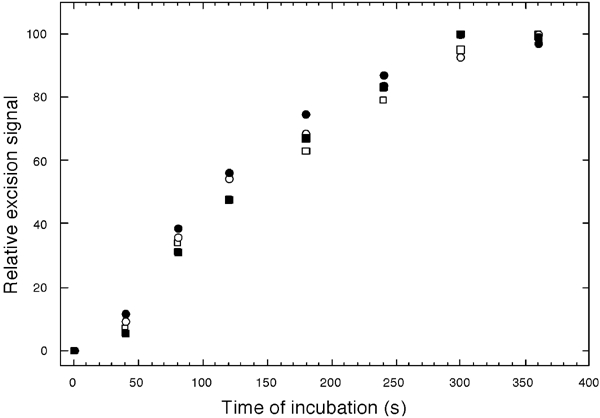
Time courses of mismatch-provoked excision. Preparation of circular substrate (E), incorporating a single biotinylated nucleotide in both paths from the (G/T) mismatch to a pre-existing nick (□, ○), and substrate (F), with streptavidin–biotin complexes at the same positions (▪, •), was as described under ‘Materials and methods'. Substrates were mixed on ice with HeLa nuclear extracts, in MMR buffer lacking exogenous dNTPs, and temperatures were immediately raised to 37°C (time zero). At the times indicated, aliquots were removed and DNA was extracted and analyzed for excision of the nicked strand nt 30–60 along the shorter (3′ to 5′) path towards the mismatch, by annealing of probe #3, gel electrophoresis, and phosphorimaging, as described under ‘Materials and methods'. Excision signals (amounts of probe radioactivity bound) are expressed relative to plateau values at 300 or 360 s. Squares and circles correspond to two independent trials.
