Figure 1.
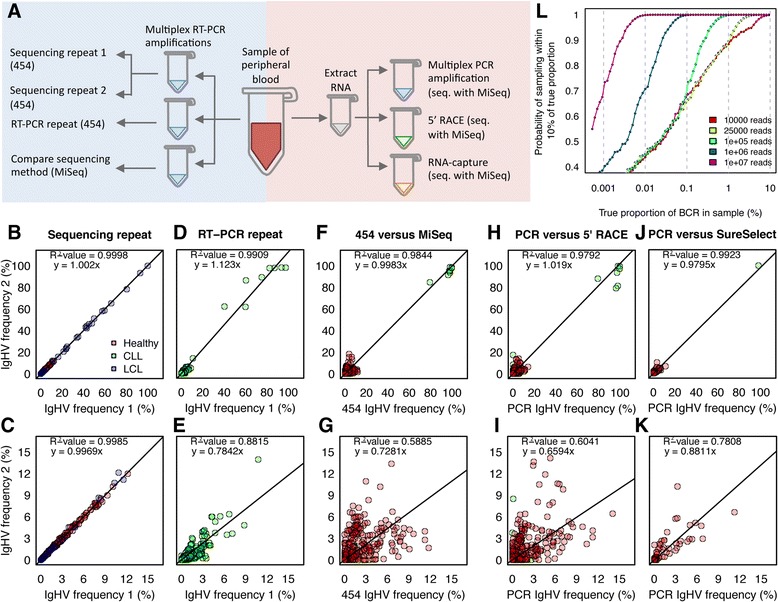
Comparing different RNA-capture and amplification methods. A) Schematic diagram of all experiments. Left side: RNA was extracted from B-cell samples, and multiplex RT-PCR performed in triplicate: sequencing repeats (re-sequencing the same PCR products), PCR repeats (independent RT-PCR of the same RNA and sequencing by MiSeq) and sequencing method comparisons (independent RT-PCR of the same RNA source and sequenced by 454 and MiSeq). Right side: RNA was extracted from B-cell samples, and 5’RACE (by MiSeq), RNA-capture (by MiSeq) were compared to PCR amplification of the same samples (using 454 sequencing). Graphs of IgHV gene-usage frequency distributions between samples were generated from B) the sequencing repeats, D) RT-PCR repeats, F) sequencing method comparisons, H) multiplex PCR versus 5’RACE (by MiSeq), J) multiplex PCR versus RNA-capture (sequenced by MiSeq). Graphs C, E, G, I and K) are IgHV gene-usage frequency distributions from only the low frequencies (<15%) respectively. Point colors are red, blue and green for healthy, LCL and CLL samples respectively. The linear regression equation and R2-values are given. L) Plot of the probability of sampling within 10% of the true of a BCR proportion with varying read depths (10,000, 25,000, 100,000, 1,000,000 and 10,000,000 reads) assuming an initial population of 50,000,000 BCR sequences after amplification.
