Figure 2.
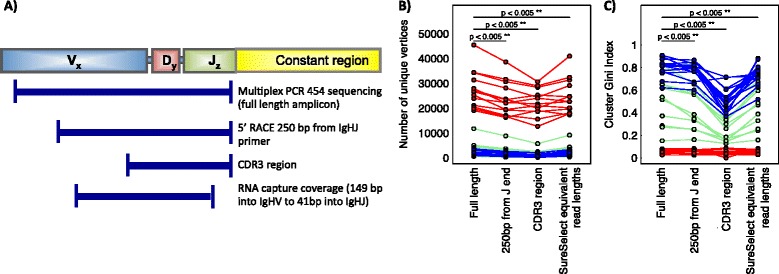
Variation of diversity measures with read-length. A) Schematic diagram showing the read-lengths from each technique aligned against the BCR gene. 454 multiplex sequencing reads were trimmed either between i) containing bases within 250 bp from the end of the IgHJ region, ii) CDR3 region, iii) or the mean region covered by reads from the RNA-capture method (149 bp from 3’end of IgHV to 41 bp from 5’end of IgHJ), and corresponding BCR networks were generated. Plots show the variation of B) number of unique sequencing reads and C) Cluster Gini Index. Point colors are red, green and blue for healthy PBMC, LCL and CLL samples respectively.
