Figure 5.
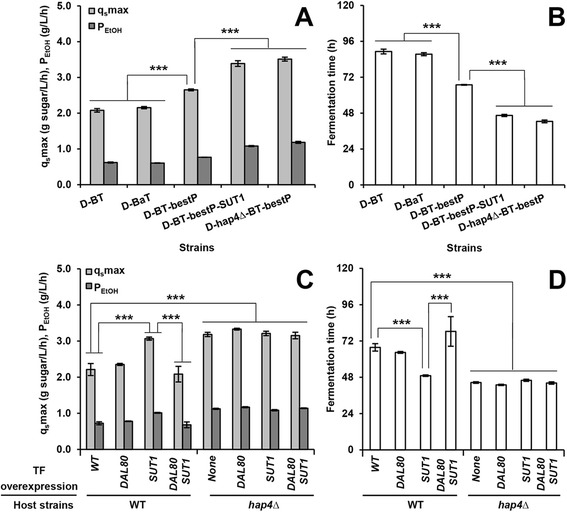
Comparisons of cellobiose fermentation using strain D452-2 with the original (P PGK1 -driven cdt-1 and gh1-1a ) and optimized (P TDH3 -driven cdt-1 and P CCW12 -driven gh1-1a ) cellobiose-utilizing pathways and transcription factor (TF) mutants. (A) Cellobiose-consumption rates and ethanol productivities with strains expressing the original and optimized cellobiose-utilization pathways, and also either overexpressing SUT1 or harboring a hap4 deletion. For details of the strains, see Additional file 4: Table S1. (B) Fermentation times of the strains in (A). (C) Comparisons of cellobiose-consumption rates and ethanol productivities with WT D452-2 or D452-2 (hap4Δ) expressing the optimized cellobiose-utilization pathway, and additionally overexpressing SUT1 and/or DAL80. (D) Fermentation times of the strains in (C), defined as the time when ethanol reached its maximum titer. In all experiments in (A–D), an initial OD600 of 20 was used. Data represent the mean and standard error of triplicate cultures on each source. Statistical analysis in (A) and (C) was performed using two-way ANOVA (with strains and fermentation rate including qsmax and PEtOH as the factors) followed by Tukey’s multiple-comparison posttest (***P < 0.001). Statistical analysis in (B) and (D) was performed using one-way ANOVA followed by Tukey’s multiple-comparison posttest (***P < 0.001).
