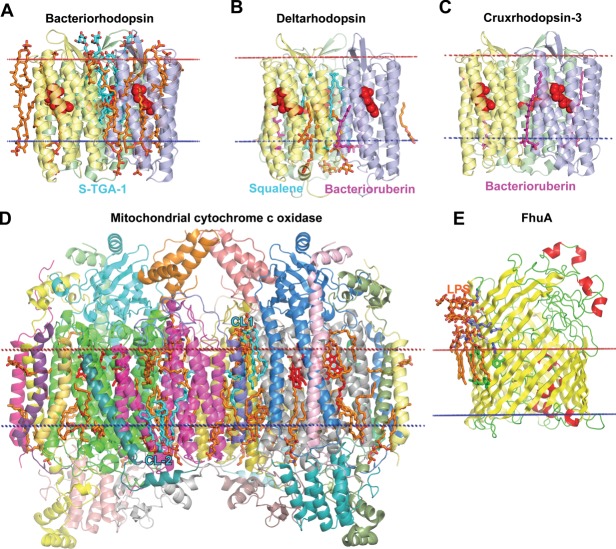
Figure 14.
Specific protein–lipid interactions. (A) Bacteriorhodopsin trimer from Halobacterium salinarium (1IW6) with co-crystallized archaeol lipids (colored orange), including S-TGA-1 sulfoglycolipid (3-sulfate-Galpβ1–6Manpα1–2Glcpα1-archeol) colored cyan. (B) Deltarhodopsin trimer from Haloterrigena thermotolerans (4FBZ) with co-crystallized lipids/detergents (colored orange), 3 squalene molecules (colored cyan) occupying intertrimer space, and 3 bacterioruberin moleculs (colored purple) located at monomer interfaces. (C) Cruxrhodopsin-3 trimer from Haloarcula vallismortis (4JR8) with 3 co-crystallized bacterioruberin moleculs (colored purple) located at monomer interfaces. (D) Dimeric mitochondrial cytochrome c oxidase from bovine heart (2DYR) co-crystallysed with 26 lipids (colored orange) including four hemes A (colored red) and four CLs (colored cyan): two CLs (CL-1) at the intermembrane side that likely participate in dimer stabilization, two CLs (CL-2) molecules at the matrix side that may participate in the proton translocation along the protein surface.313 In the latter case CL in each monomer interacts with basic residues from subunits 3 and 7A1. (E) Ferrix hydroxamate uptake receptor FhuA from E. coli (colored by secondary structure: yellow β-strands, red helices, and green loops) in complex with LPS (colored orange) (1QFG). Lipid A of LPS forms specific interactions with a number of basic (shown as blue sticks), polar (white sticks) and aromatic residues (green sticks) of FhuA.