Figure 2.
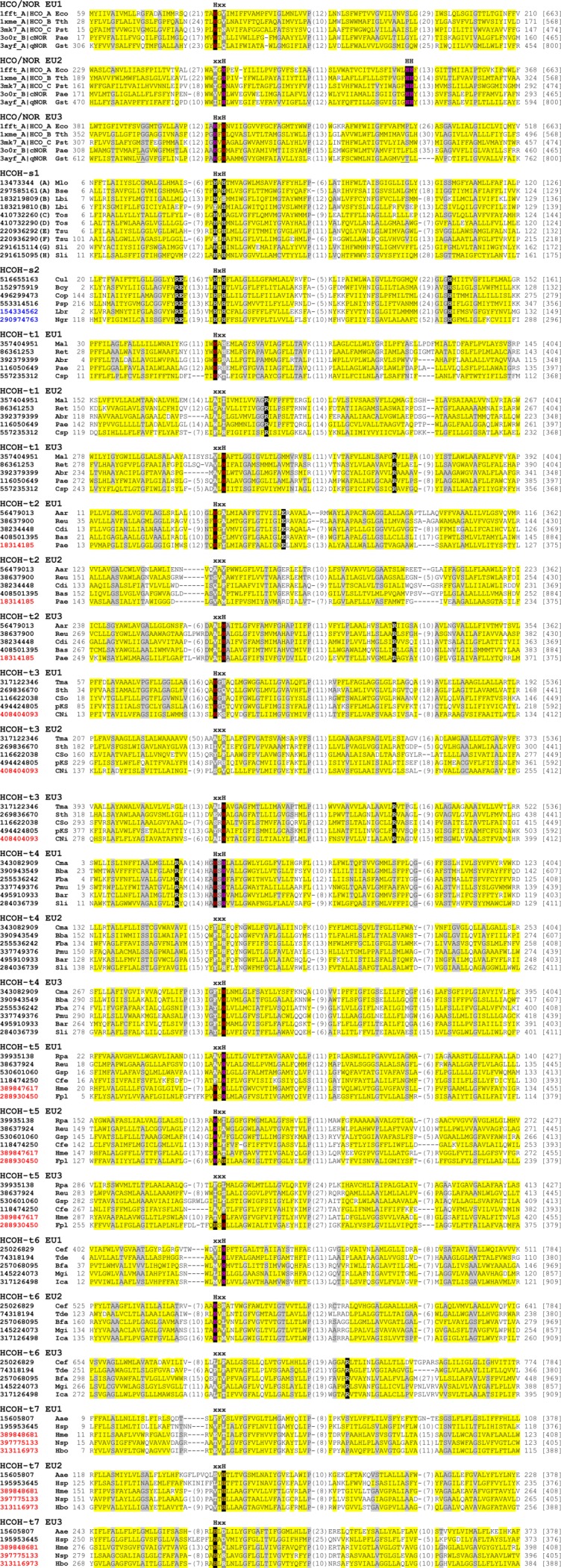
Multiple sequence alignment of EUs of HCOs/NORs and HCOH proteins. For HCOs/NORs, the sequences are denoted by their pdb id and chain id, followed by type name. For HCOH proteins, each sequence is denoted by its NCBI gene identification (gi) number. The cluster type for HCOH-s1 is shown in parentheses after gi numbers. Starting and ending residues numbers are shown before and after the sequences, respectively. Protein lengths are shown in brackets at the end. Positions with conserved motifs in the second TMHs of these EUs are marked with HxH, xxH, Hxx, or xxx. The conserved HH motif in the second EU of HCOs/NORs is also marked. Conserved histidines in these positions are in black background. For three-EU proteins, conserved histidines at the interface between EU1 and EU3 are colored red, those at the interface between EU2 and EU3 are colored orange, and those at the interface between EU1 and EU2 are colored magenta. Other family-specific conserved residues are also highlighted in black background. Non-charged residues in positions with mainly hydrophobic residues are shaded in yellow. Small residues (G,A,S,C,T,P, and V) in positions with mainly small residues are shaded in grey. Insertion regions are replaced by the number of inserted residues in parentheses or omitted in between underscored letters. Three-letter species name abbreviations shown before the residue starting numbers are as follows: Aae, Aquifex aeolicus; Aar, Aromatoleum aromaticum; Abr, Azospirillum brasilense; Bcy, Bacillus cytotoxicus; Bse, Bacillus selenitireducens; Bba, Belliella baltica; Bas, Bifidobacterium asteroides; Bar, Bizionia argentinensis; Bfa, Brachybacterium faecium; Cfe, Campylobacter fetus; Csp, Campylobacter sp.; CNi, Candidatus Nitrososphaera gargensis; CSo, Candidatus Solibacter usitatus; Cop, Coprobacillus; Cdi, Corynebacterium diphtheriae; Cef, Corynebacterium efficiens; Cul, Corynebacterium ulceribovis; Cma, Cyclobacterium marinum; Eco, Escherichia coli; Fpl, Ferroglobus placidus; Fba, Flavobacteriaceae bacterium; Gsp, Geobacillus sp.; Gst, Geobacillus stearothermophilus; Hme, Haloferax mediterranei; Hbo, Halogeometricum borinquense; Hsp, Hydrogenobaculum sp.; Ica, Intrasporangium calvum; Lbr, Leishmania braziliensis; Lbi, Leptospira biflexa; Mlo, Mesorhizobium loti; Mal, Methylomicrobium alcaliphilum; Mgi, Mycobacterium gilvum; Ngr, Naegleria gruberi; Nsp, Natrinema sp.; pKS, planctomycete KSU-1; Pmu, Paenibacillus mucilaginosus; Psp, Peptoniphilus sp.; Pae, Pseudomonas aeruginosa; Pst, Pseudomonas stutzeri; Pae, Pyrobaculum aerophilum; Reu, Ralstonia eutropha; Ret, Rhizobium etli; Rpa, Rhodopseudomonas palustris; Sli, Sideroxydans lithotrophicus; Sth, Sphaerobacter thermophilus; Sli, Spirosoma linguale; Tma, Thermaerobacter marianensis; Tos, Thermus oshimai; Tth, Thermus thermophilus; Tsu, Thioalkalivibrio sulfidophilus; Tde, Thiobacillus denitrificans. Gi numbers of bacterial, archaeal, and eukaryotic proteins are in black, red, and blue colors, respectively.
