Abstract
Sortilin is a multifunctional receptor involved in sorting and apoptosis. We have previously reported a 2.0-Å structure of the Vps10 ectodomain in complex with one of its ligands, the tridecapeptide neurotensin. Here we set out to further characterize the structural properties of sortilin and its interaction with neurotensin. To this end, we have determined a new 2.7 Å structure using a crystal grown with a 10-fold increased concentration of neurotensin. Here a second peptide fragment was observed within the Vps10 β-propeller, which may in principle either represent a second molecule of neurotensin or the N-terminal part of the molecule bound at the previously identified binding site. However, in vitro binding experiments strongly favor the latter hypothesis. Neurotensin thus appears to bind with a 1:1 stoichiometry, and whereas the N-terminus does not bind on its own, it enhances the affinity in context of full-length neurotensin. We conclude that the N-terminus of neurotensin probably functions as an affinity enhancer for binding to sortilin by engaging the second binding site. Crystal packing differs partly from the previous structure, which may be due to variations in the degree and pattern of glycosylations. Consequently, a notable hydrophobic loop, not modeled previously, could now be traced. A computational analysis suggests that this and a neighboring loop may insert into the membrane and thus restrain movement of the Vps10 domain. We have, furthermore, mapped all N-linked glycosylations of CHO-expressed human sortilin by mass spectrometry and find that their locations are compatible with membrane insertion of the hydrophobic loops.
Keywords: crystal structure, sortilin, neurotensin receptor 3, neurotensin, Vps10 domain (Vps10p domain), β-propeller, hydrophobic loop, membrane imbedded loop, N-linked glycosylations
Introduction
Sortilin, also known as neurotensin receptor 3 (NTS3 or NTR3), is a multifunctional receptor belonging to the Vps10 domain family, or sortilins family, which in mammals also includes SorLA and the mutually homologues SorCS1, SorCS2, and SorCS3.1 It predominates in intracellular compartments and participates in endocytosis as well as intracellular trafficking, for example, retrograde transport and cycling between endosomes and the trans-Golgi network.1 Sortilin is, however, also found at the cell surface and is capable of triggering neuronal apoptosis by forming a death-signaling trimeric complex with proneurotrophins and the p75NTR receptor.2,3 Sortilin is synthesized as a proprotein with a 44 residues long propeptide, which plays an important role in facilitating transport of the proreceptor through the biosynthetic pathway, until it is cleaved off in the trans-Golgi network.4,5 Neurotensin (NT) is a neuropeptide of 13 amino acids, which may induce a number of different biological activities including hypothermia, analgesia, hypotension, and cell proliferation involving a variety of neuronal systems including dopaminergic and serotonergic circuits.6 The physiological role of binding of NT to sortilin is still unclear, but it has been suggested that sortilin, alone or in complex with the G protein-coupled NT receptor-1, may convey NT signaling.7–9 Notably, both NT and the released sortilin propeptide (Sort-pro) bind mature sortilin with high affinity and can effectively compete for binding with each other, as well as with almost all other known ligands.2–4,10 We have previously reported the structure of the Vps10 ectodomain of human sortilin (sSortilin) in complex with NT (PDB accession code: 3F6K).11 This structure revealed that the Vps10 “domain” is composed of three individual domains; a large 10-bladed Asp-box β-propeller domain and two small cysteine rich domains named 10CC-a and 10CC-b, and that it contains at least two N-linked glycosylations. NT was found to bind within the tunnel of the β-propeller domain via its three C-terminal residues, YIL,11 and we have recently shown that also small molecule inhibitors of NT binding occupies this YIL binding site.12,13
Here we have set out to further study the structural properties of sortilin and the binding of NT using a broad biophysical approach. The data provided here suggest that NT is completely contained inside the tunnel of the β-propeller, firmly bound by its C-terminus in the previously reported binding site, and further attached by its N-terminal part at a low affinity site on the opposite side of the tunnel. In addition, our data suggest possible roles for the hydrophobic loops and glycosylations of the β-propeller domain.
Results
Overall structure and crystal packing
We have determined a structure of sSortilin in complex with NT (sSort:NTHigh) at 2.7-Å resolution using a crystal grown with a 10-fold increased concentration of NT as compared with the previously determined structure of sSort:NT (PDB: 3F6K; sSort:NTLow). Overall, the two structures are almost identical, which is reflected in a very low pairwise root mean square deviation of 0.44 Å for 603 Cα positions. However, a few interesting differences exist; a second peptide binding site is occupied in sSort:NTHigh, as discussed later, and additional differences exist in the glycosylations and hydrophobic loops. These additional differences seem to relate to differences in crystal packing. The new structure represents a less compact crystal form than sSort:NTLow [Fig. 1(a,b)], although some packing interactions are shared between the two forms [Fig. 1(c,d)]. Interestingly, the far majority of crystals grown at low concentration of NT or with small molecule inhibitors used in previous studies, adopt the same crystal form as sSort:NTHigh, even though they were grown under similar and sometimes even identical conditions to those used for the sSort:NTLow native crystal. Hence, it is neither the type and concentration of the peptide ligand nor the differences in the composition of the crystallization solution that dictates which of the two crystal forms is adopted. One possibility is that the crystals adapt to variation in glycosylation site occupancy (also known as glycosylation macroheterogeneity), which is a common phenomenon for proteins purified from mammalian cells including CHO cells.14 Thus, whereas there in accordance with the mass spectrometry data (see later) is weak electron density for a single N-acetyl glucosamine residue at Asn129 in the new structure, no glycosylation seems present here in sSort:NTLow. Indeed, modeling of a glycosylation at Asn129 suggests that a glycosylation here is incompatible with the crystal packing of sSort:NTLow (Supporting Information Fig. 1).
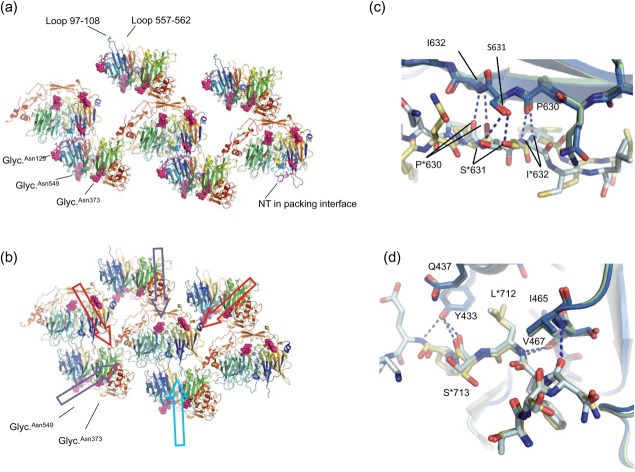
Figure 1.
Crystal packing. (a) Packing of sSort:NTHigh. sSortilin is shown in cartoon colored rainbow (N-terminal blue, C-terminal red). The glycosylations and NT (hardly visible) are colored pink and shown in space-fill and sticks, respectively. The NT molecule trapped in a crystal packing interface is labeled along with the glycosylations and hydrophobic loops. (b) Packing of sSort:NTLow. Some interactions are shared with sSort:NTHigh (violet arrows). However, one interaction face, which includes the hydrophobic loops, is very different (light blue arrow) leading to more compact packing with additional interaction faces (red arrows), which no doubt underlies the higher diffraction potential. (c) Common packing interactions across the β-sheet of 10CC-a (this is the interaction face that is indicated by the top violet arrow in panel b). sSort:NTLow is light and dark blue and sSort:NTHigh is yellow and green. This interface is mainly formed by polar interactions. Hydrogen bonds are indicated by dashed lines and selected residues are labeled (an asterisk is used for the symmetry related molecule). (d) Common packing interactions between a stretch of C-terminal residues and loops of blade 8 (violet arrow at bottom left in panel b). This interface is formed both by polar interactions involving the main chain of the C-terminal stretch and by burial of Leu713 in a hydrophobic pocket formed by the side chains of Tyr433, Gln437, Ile465, and Val467.
Hydrophobic loops
Due to the differences in crystal packing interactions, the hydrophobic loop 97–108 at blade 1 of the β-propeller and the neighboring loop 557–563 at blade 10, which is hydrophobic at the tip (F559, L560), could be modeled to different extents in the two structures. Loop 557–563 is partly disordered in the new structure, whereas it is well ordered in sSort:NTLow, presumably because it is involved in crystal packing in this crystal form. In contrast, loop 97–108 is much better ordered in the new structure due to formation of packing interactions in the β-propeller tunnel of a symmetry related molecule. It could, therefore, for the first time be modeled in its entirety and presents itself as the most protruding loop of all the loops of the sSort:NTHigh structure [Fig. 2(a,b)]. We have previously reported a multiple sequence alignment of sSortilin from eight species with pairwise identities ranging from 60 to 95%.11 A closer examination of this alignment shows that the hydrophobic loop at blade 1 is markedly better conserved than the loop at blade 10 [Fig. 2(c)], but not more than moderately conserved as compared to the overall conservation of the Vps10 domain. Yet, the hydrophobic positions are fully conserved as hydrophobic in both loops. Interestingly, a multiple alignment of the five human sortilins (Supporting Information Fig. 2) shows that both loops are markedly less hydrophobic in SorLA and SorCS1–3 than in sortilin [Fig. 2(d)]. A computational prediction of putative membrane inserting loops suggests that both residues 100–106 (using sSort:NTHigh as input) and residues 558–561 (using sSort:NTLow as input) may be membrane-imbedded segments [Fig. 2(a)].
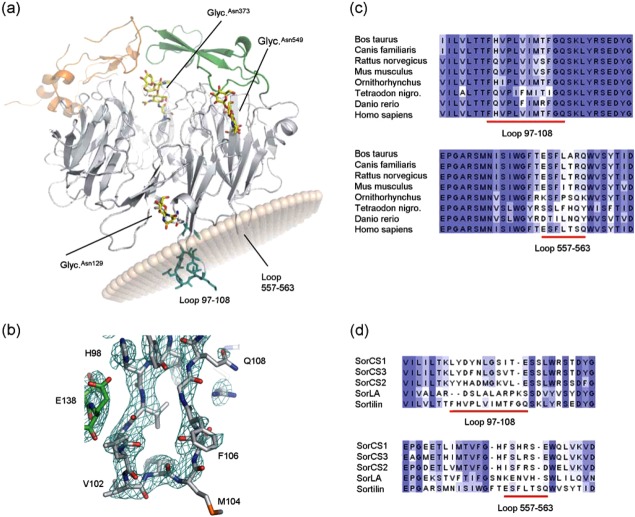
Figure 2.
Hydrophobic loops. (a) Cartoon of sSort:NTHigh, the propeller is gray, 10CC-a is green, 10CC-b is orange, glycosylations are shown in yellow sticks and the hydrophobic residues of loop 97–108 are shown in turquoise. The position of the membrane was predicted using the PPM server (light brown transparent spheres). However, the tilt angle is somewhat unreliably predicted due to missing residues. A similar analysis of sSort:NTLow shows that also loop 557–563 is likely to insert into the membrane (not shown). (b) Close-up of loop 97–108. The 2Fo-1Fc electron density map is shown at the 1σ level in turquoise wire mesh. Part of a symmetry related molecule that interacts with loop 97–108 can also be seen in this view (green). (c) Multiple alignments of sSortilin and seven vertebrate homologues (colored in shades of blue according to conservation). The regions around loop 97–108 and loop 557–563 are shown. (d) Multiple alignments of the Vps10 domains of all human Vps10 proteins. The regions around loop 97–108 and loop 557–563 are shown (the full alignment can be seen in Supporting Information Fig. 2). Both loops appear to be longer in sortilin, loop 97–108 is markedly more hydrophobic, and there are two rather than one hydrophobic residues at the tip of loop 557–563.
Ligand binding
In the sSort:NTHigh structure, we observe no changes in the previously identified binding site at blade 6, which we will now refer to as binding Site 1, but additional electron density has appeared at a second binding site (Site 2) on the opposite side of the tunnel at blade 1 (Fig. 3). This density suggests the presence of a pentapeptide forming a short β-strand interacting with Strand 1 of Blade 1, but the side chains are not unambiguously defined [Fig. 3(c)]. The electron density for the peptide in Site 2 adjoins its symmetry related counterpart, suggesting that one molecule of NT may span two molecules of sSortilin. If this is the case, it would be incorrect to process the data and refine the structure in the C2 space group. However, when refined in space group P1, the final electron density map was in best agreement with one peptide in Site 2 per one molecule of sSortilin, and no clear improvement was observed with respect to the definition of side chain electron density (not shown). We are confident that the peptide observed in Site 2 is not an artifact from purification, since we found no peptide here in complexes formed at low NT concentrations using the same preparation of sSortilin. It follows that it either represents a second molecule of NT, or the N-terminal part of the molecule bound in Site 1 by its C-terminus. If the latter is true, the middle part of the peptide would be disordered, as we see no peptide electron density in the middle of the tunnel. To further examine this, we first tested the ability of full-length NT and NT8–13 to compete for binding with GST-PYIL (PYIL corresponds to NT10–13) in a surface plasmon resonance (SPR) experiment. A sixfold reduction in the IC50 values obtained shows that the N-terminal residues of NT are important for efficient competition [Fig. 4(a)]. Next we examined the affinity of sSortilin for NT1–8, NT8–13, and full-length NT by isothermal titration calorimetry (ITC). For NT1–8, no binding was observed, that is, no heat release/absorption could be measured under the experimental conditions (data not shown), whereas we for full-length NT and NT8–13 obtained Kd = 85 nM and Kd = 337 nM with a stoichiometry of 1, respectively [Fig. 4(b,c)]. We conclude that the N-terminal part of NT contains elements, which increase the affinity for sortilin, but are insufficient for binding with high affinity on its own. In the final structure we have therefore chosen to model the pentapeptide observed in Site 2 as the N-terminal part of the NT molecule that is bound in Site 1 via its C-terminus. Specifically, we have modeled the peptide as NT1–5, as shifting the sequence to for example, NT2–6 would not leave enough residues to connect the N- and C-terminal segments. A third peptide binding site was also identified, but we interpret this as an artifact, since it is found at a nonconserved patch on the outside of the propeller and is part of a crystal packing interface [Fig. 1(a)].
Figure 3.
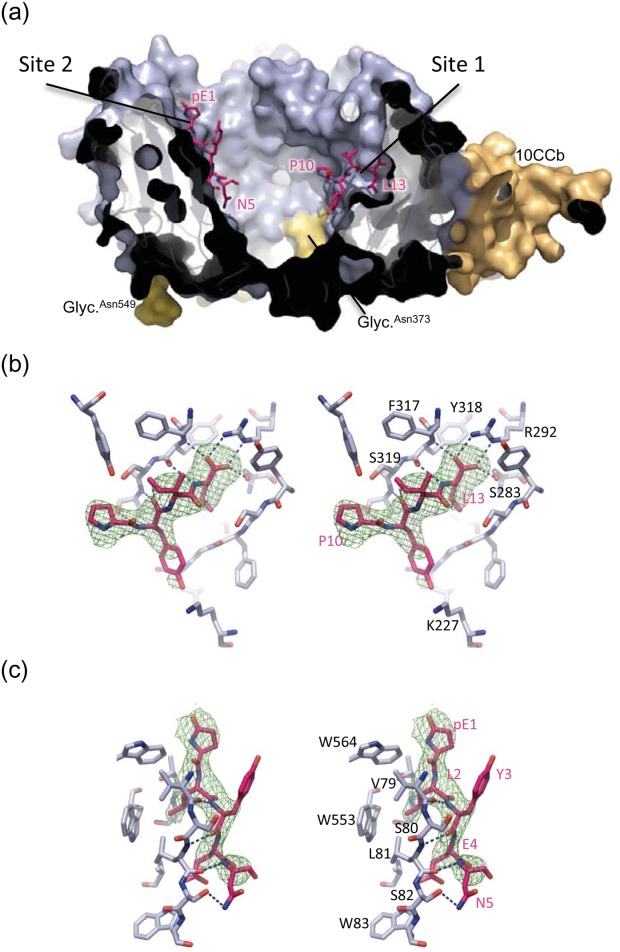
Neurotensin binding sites. (a) Cross-section of sSort:NTHigh shown in surface representation. The propeller domain is blue, 10CC-a is green (hardly visible in this view), 10CC-b is orange, the glycosylations are yellow and labelled, and NT is shown in pink sticks. (b) Stereo image of NT in binding site 1 of sSort:NTHigh. Sortilin residues within 4 Å of the bound NT are shown. Dashed lines indicate positions of hydrogen bonds, i.e., distances shorter than 3.2 Å, and the green wire mesh represents the SA-omit map of NT contoured at 3σ. Labels of selected residues of sortilin and NT are shown in black and pink respectively (c) Stereo image of NT in binding site 2 of sSort:NTHigh. Hydrogens bonds, SA-omit map and labels are shown as in panel b.
Figure 4.
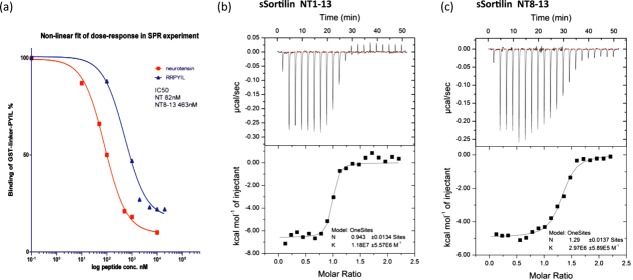
Peptide binding experiments. (a) Competition of NT and NT8–13 with GST-Linker-PYIL measured by SPR. Derived IC50 values are indicated. (b) ITC measurement of NT binding to sSortilin. (c) ITC measurement of NT8–13 binding to sSortilin.
Glycosylation pattern
To further expand on the structural characterization of sortilin, we next mapped all N-linked glycosylations by mass spectrometry [Supporting Information Table I and Fig. 5(a)]. Most are on the bottom face (Asn373, Asn549) or rim (Asn65, Asn129) of the β-propeller, but one is also found at the 10CC-b domain (Asn651) and one at the top face (Asn241). It should be mentioned that the peptide containing Asn549 for unknown reasons was not observed in the MS data. However, both sSort:NT structures clearly show that it is present. To test the function of the glycosylations, we carried out a chase experiment on CHO cells grown with tunicamycin, a known inhibitor of N-linked glycosylation [Fig. 5(b)]. This experiment showed that under these conditions, sSortilin is secreted at a strongly reduced rate, indicating that at least some of the glycosylations are required for proper folding and/or intracellular transport of the receptor.
Table I.
Diffraction Data and Structure Refinement Statistics
| sSort-NTHigh | |
|---|---|
| Beamline | I911-5, MAX-lab |
| Data statistics | |
| Wavelength (Å) | 0.90736 |
| Resolution limit (Å) | 21.0–2.66 (2.76–2.66) |
| Space group | C2 |
| Unit-cell | |
| a | 162.01 Å |
| b | 78.65 Å |
| c | 110.98 Å |
| β | 126.62° |
| Rmerge (I) | 5.4 (74.4) |
| Completeness | 99.2 (99.0) |
| I/sigma | 16.8 (2.23) |
| Wilson B-factor | 68.1 |
| Refinement statistics | |
| R-factor | 0.166 (0.254) |
| Rfree | 0.212 (0.293) |
| Number of atoms in model | 5518 |
| sSortilin and NT | 5305 |
| Ligands | 105 |
| Water | 108 |
| Mean B-factor (Å2) | 86.3 |
| sSortilin | 86.4 |
| NT in binding site 1+2 | 118.1 |
| NT at packing interface | 121.3 |
| Glycosylations | 110.9 |
| Solvent (water, glycerol) | 70.6 |
| Geometry | |
| RMSD bond lengths | 0.004 Å |
| RMSD bond angles | 0.74° |
| Phi-Psi distribution | |
| Most favored | 95.0% |
| Allowed | 4.7% |
| Dissallowed | 0.3% |
Numbers in parentheses are for the highest resolution shell. The structure belongs to the same space group as the previously reported sSort:NTLow structure (PDB: 3F6K), but the cell dimensions are substantially different (cell parameters for 3F6K: a = 145.76, b = 74.53, c = 108.33, β = 131.87).
Figure 5.
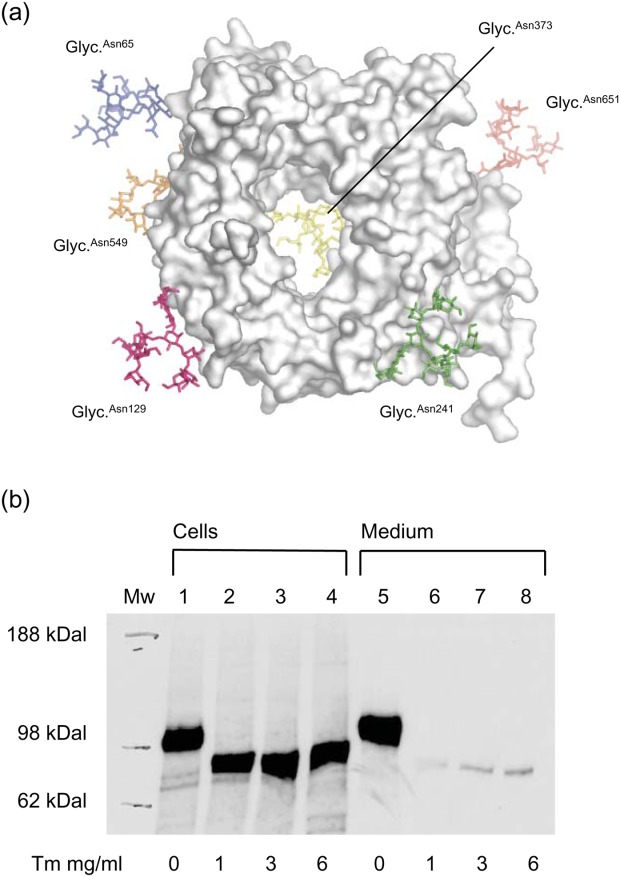
Glycosylations. (a) sSort:NTLow is shown in a top face view with N-linked glycosylations (sticks) added in silico at sites determined by MS to be glycosylated. They are colored blue at Asn65, light blue at Asn129, green at Asn241, yellow at Asn373, orange at Asn549, and red at Asn651. For reference, the hydrophobic loops are also colored; loop 97–108 is turquoise and loop 557–563 is purple. (b) Tunicamycin (Tm) effect on sSortilin secretion. Lanes 1–4 show sSortilin content of lysed cells and lanes 5–8 show sSortilin content in the growth media after addition of tunicamycin in various concentrations, as indicated below each lane.
Discussion
Differences in crystal packing
Due to a pronounced difference in crystal packing, the new sSort:NT structure presented here could not be determined to as high a resolution as the previously determined structure. The reason for these packing differences is unknown, but they may be caused by variable glycosylation site occupancy at Asn129. We have shown that the N-linked glycosylations are important for folding and/or transport of sortilin, but it is possible that some of the six glycosylations are dispensable. If it is possible to mutate Asn129, for example, to alanine, without compromising secretion, it may thus prove possible to reproducibly obtain well diffracting crystals of future sSortilin:ligand complexes.
Putative peripheral membrane anchors
The long protruding hydrophobic loop at Blade 1 and the neighboring loop with a hydrophobic tip at Blade 10 expose a large number of hydrophobic residues to the solvent. This is energetically highly unfavorable and yet they are conserved as hydrophobic in all species. We therefore believe that they serve important functions. Furthermore, it is noteworthy that they protrude from the top face of the β-propeller, since the functionally important loops of all other known structures of Asp-box β-propellers are all found on this face.15 Notably, any function of the loops is probably specific to sortilin rather than applying to the Vps10-D family in general, since they are somewhat shorter and less hydrophobic in SorLA and indeed much less hydrophobic in SorCS1–3. We have previously speculated that they could be involved in membrane anchoring in conjunction with the single transmembrane helix found C-terminally of the Vps10 domain, or in interaction with hydrophobic patches on protein ligands or receptor partners. Hydropathy plots did indeed identify the loop at Blade 1 as a putative transmembrane segment, but it is highly unlikely that it tightly fixes the top face of the β-propeller against the membrane surface, as the top face is not distinctly basic, and as ligands are probably unable to enter the tunnel from the very narrow entrance found at the bottom face.11 It is however possible that the loops serve as membrane anchors, which reduce the freedom of movement of the domain without clamping it tightly to the membrane surface. This hypothesis is supported by a computational analysis, which suggests that both hydrophobic loops may function as peripheral membrane anchors. It is also noteworthy that the only glycosylated residue on the top face is Asn241, which is located on a long flexible loop >35 Å away from the hydrophobic loops. Glycosylation of the protein is thus unlikely to impede membrane insertion of the hydrophobic loops. Furthermore, anchoring via hydrophobic loops may also explain why tumor necrosis factor α-converting enzyme (TACE)-mediated ectodomain shedding is much slower for sortilin than for any of the other human Vps10-D receptors except SorCS2,16 since such anchoring may hamper the release of ectodomains from the membrane. If indeed the hydrophobic loops serve as membrane anchors, they could for example be important for proper alignment of the domain for interaction with its cognate receptor partners and/or for controlling access to the ligand binding sites within the β-propeller tunnel.
A second neurotensin binding site
The peptide observed in Site 2 in the sSort:NTHigh structure may in principle either represent a second molecule of NT, or the N-terminal part of the molecule bound to site 1 by its C-terminus. However, we strongly favor the latter hypothesis. Early experiments on binding of NT to sortilin thus suggested a 1:1 stoichiometry,17 which is also in agreement with the ITC data presented here. In addition, neither NT1–8 nor C-terminally amidated NT8–13 can displace full-length NT in binding competition experiments,11,18 which strongly disfavors the hypothesis that two peptides are bound, as binding at Site 2 is clearly not dependent on the presence of a free C-terminus. Furthermore, we find that the ability of NT to compete for binding with GST-PYIL is reduced sixfold in an SPR experiment, and that the affinity of binding, as determined by ITC, is reduced fourfold when the 7 N-terminal most residues are removed, implying that these residues are involved in binding. Yet, we were unable to detect any binding of the C-terminally truncated NT1–8 peptide using ITC. We, therefore, conclude that the N-terminal residues enhance the affinity in context of full-length NT, but are insufficient to bind on their own with appreciable affinity. A role of the N-terminal part in acting as an affinity enhancer in context of full-length NT is furthermore supported by the observation that arginylation of Glu4, a naturally occurring NT modification, leads to a threefold increase in binding affinity to sortilin.19 We therefore conclude that NT is most likely bound as follows: The C-terminus is bound in Site 1 with high affinity, the middle part is disordered in the tunnel, and the N-terminus is bound in Site 2 with low affinity and probably low specificity, as indicated by the absence of well-defined side chain electron density for bound NT. Notably, binding of NT in Site 2 may not only enhance the affinity of binding but may also partly underlie the striking ability of NT to compete for binding with most other ligands, as it would cause severe steric hindrance of access to binding sites within the tunnel of the β-propeller domain. Binding Site 2 is in likeness with Site 1 found in a highly conserved patch, indicating that it probably serves an important function (Supporting Information Fig. 3). This may solely relate to its apparent role as an auxiliary binding site for NT. However, it is possible that binding Site 2 is also involved in binding of other ligands as either a primary or auxiliary binding site. Additional binding experiments and structures of more sSortilin:ligand complexes may help to resolve this.
Materials and Methods
Protein and peptide preparation
The ectodomain of human sortilin, sSortilin, including the signal peptide (amino acids −33 to 725), was expressed in CHO-K1 cells and purified in one single step by RAP affinity chromatography as described previously.2,20 NT was purchased from Sigma and was more than 95% pure. Prior to crystallization, the purified protein was dialyzed into a buffer containing 50 mM Tris-HCl pH 7.9 and 150 mM NaCl and concentrated to 4.5–5.5 mg/mL, as determined by Bradford assay, using either Centricon (Millipore Corp.) or Vivaspin (Sartorius Ltd.) concentrators. Protein samples prepared in this way are very stable and can be stored at −20°C for at least 5 years.
Crystallization and structure determination
Mixing with NT was done between a few minutes and an hour before crystallization experiments were set up using an sSort:NT ratio of about 1:15 (10-fold increased concentration of NT as compared with the previous sSort:NTLow structure). Crystallization experiments were performed at 19°C by sitting-drop vapor diffusion using 2 μL + 2 μL of protein and reservoir solution consisting of 20% PEG 6000, 300 mM sodium malonate pH 6.7, 3% glycerol and 100 mM Tris-HCl pH 7.9. Cryoprotection was achieved by dehydration: The reservoir was replaced by a buffer containing the same amounts of PEG and salt, but with the glycerol content increased to 12% and resealed to equilibrate over night before flash-freezing the crystals in liquid nitrogen. The sSort:NTHigh dataset was collected at Max-lab beamline 911-5 from a crystal with longest dimension of 250 μm. The data were processed using XDS21 (Table I). Phasing was achieved by molecular replacement using PDB: 3F6K stripped from carbohydrate, solvent and ligands as input for Molrep.22 Refinement was carried out by alternating cycles of maximum likelihood refinement with PHENIX refine23 using translation libration screw (TLS) restraints (Table I) and manual rebuilding in O.24
Sequence and structural analysis
Multiple sequence alignment of the Vps10 domains of human sortilin (UniProtKB/Swiss-Prot Q99523: Residues 45–716), human SorLA (Q92763: 82–761), human SorCS1 (Q8WY21:111–801), human SorCS2 (Q96PQ0: 51–784) and human SorCS3 (Q9UPU3: 134–825) was carried out using ClustalW2.25 The alignment was then edited guided by the sSort:NTLow structure using Jalview 2.4.26 Prediction of membrane interacting loops was carried out using the PPM server (http://opm.phar.umich.edu/server.php).27 All figures of the structures were made using PyMol.28
Surface plasmon resonance experiments
NT and NT8–13 were purchased from Sigma-Aldrich. SPR competition experiments were carried out on a Biacore 3000 instrument against 100 nM GST-linker-Pro-Tyr-Ile-Leu (linker sequence; QDRLDAPPPPAAPL) using conditions and procedures as previously reported.2 Each measurement of GST-PYIL with peptide was corrected for the background with peptide alone and for every second measurement, GST-PYIL was measured alone to correct the maximal signal for degradation of immobilised sSortilin. Percentage inhibition was calculated as the response units of mixed peptide and GST-linker-PYIL subtracted the response units for the peptide alone divided by the response units of GST-linker-PYIL alone. Data were processed in GraphPad Prism 5 as a least squares nonlinear fit.
Isothermal titration calorimetry experiments
Binding of NT (Sigma), NT1–8 (American Peptide Company, Sunnyvale, CA) and NT8–13 (Sigma) to sSortilin was measured on a MicroCal iTC200 isothermal titration calorimeter (MicroCal Inc., Northampton, MA). sSortilin for ITC was prepared as described above and then dialyzed against PBS buffer, pH 7.4. Peptides were dissolved in PBS buffer, and to ensure accurate measurements, concentration of protein and peptides were determined by total amino acid analysis as described previously.29 All solutions were filtered, degassed, and equilibrated to the corresponding temperature before each experiment. In a typical ITC experiment, 0.3 mL sSortilin (28 μM) was titrated at 30°C with 300 μM NT1–8 or NT8–13 in 20 steps of 2 μL (first step, 0.4 μL). Time between injections was set to 150 s and the syringe mixing speed was set at 1000 rpm. Heat of dilution was measured by injecting the ligand into PBS buffer and subtracted from the heat of reaction to obtain effective heat of binding. Finally, the binding stoichiometry (n) and equilibrium dissociation constant (Kd) for the binding processes were determined by analyzing the titration data using the ORIGIN software (OriginLab Corporation, Northhampton, MA).
Analysis of the glycosylation pattern using mass spectrometry
For identification of glycosylation sites, sSortilin was solubilized in 0.2M ammonium bicarbonate (pH 7.4) containing 10 mM dithioerythritol and incubated for 45 min at 56˚C. Free cysteines were then alkylated by addition of iodoacetamide to 50 mM and incubation for 30 min in the dark at room temperature. After the reduction and alkylation, H218O was added to a final concentration of 50% together with 4.5 mUnits PNGase F and the sample was incubated overnight at 37°C. Then, the sample was dried in a vacuum centrifuge and resuspended in 50 mM ammonium bicarbonate (pH 8.0) and incubated with trypsin (enzyme-to-substrate ratio 1:30 w/w) at 37°C overnight. Tryptic peptides were separated by reverse-phase HPLC and analyzed by MALDI-TOF-MS. De-glycosylation of Asn residues (leading to the formation of aspartic acid) in the presence of equal amounts of H216O and H218O will result in 50% incorporation of a 16O and 18O atom, respectively, in all peptides that contained a glycosylated Asn. Hence, a characteristic isotopic pattern with mass increments of 1 and 3 Da can be observed by MS of these peptides. Modeling of oligo-mannose glycosylations onto the identified glycosylation sites of sSort:NTLow was done using the GlyProt server (http://www.glycosciences.de/modeling/glyprot/php/main.php).30
Analysis of the effect on secretion of inhibiting glycosylation
For the secretion experiment, transfected CHO-K1 cells expressing sSortilin were cultured in Nunclone Multidishes (Nunc, Roskilde, Denmark). At ∼80% confluency, the cells were washed twice in Tris/balanced salt solution pH 7.5 (BSS) and incubated for 30 min in modified Eagle's medium without cysteine and methionine (Sigma Chemical Co., St Louis, MO) but supplemented with tunicamycin to 0, 1, 3, or 6 mg/mL. Excess medium was removed and 1 mL of the same medium supplemented with tunicamycin as before and 200 mCi of L-[35S]cysteine and l-[35S]methionine per milliliter of medium (Pro-mix; Amersham International, Buckinghamshire, UK) was then added to the wells. At the end of the labeling period (4–5 h), the medium was harvested and the cells were washed twice with BSS (4°C) and subsequently lysed for 10 min at 4°C in 1% Triton X-100, 20 mM Tris-HCl pH 8.0, 10 mM EDTA, 150 mM NaCl, supplemented with a proteinase inhibitor cocktail (CompleteMini; Boehringer, Mannheim, Germany). The lysed cells were spun down and CHO medium was added to the supernatant to 1 mL final volume of lysate. Each sample, either lysate or supernatant medium, was incubated with 50 μL Gammabind G-sepharose at 4°C for 2 h, after which the beads were removed. Next, a preincubated (2 h at 4°C) mixture of 75 μL Gammabind G-sepharose (Pharmacia), 500 mL CHO-medium, and 5 mL anti-sortilin Ig was added to the solution. The beads were spun down and washed five times in BSS with 0.05% tween. Protein was released from the beads by resuspension in 100 mL reducing sample buffer (10 mM DTT, 2.5% SDS) for 3 min at 95°C. Totally, 30 mL of this solution was applied to poly-acrylamide gel electrophoresis and autoradiographed.
Acknowledgments
Help from the staff at the MAX-Lab synchrotron is gratefully acknowledged.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- 1.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors—regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9:899–909. doi: 10.1038/nrn2516. [DOI] [PubMed] [Google Scholar]
- 2.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, Hempstead BL, Petersen CM. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 3.Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, Kermani P, Torkin R, Chen ZY, Lee FS, Kraemer RT, Nykjaer A, Hempstead BL. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munck Petersen C, Nielsen MS, Jacobsen C, Tauris J, Jacobsen L, Gliemann J, Moestrup SK, Madsen P. Propeptide cleavage conditions sortilin/neurotensin receptor-3 for ligand binding. EMBO J. 1999;18:595–604. doi: 10.1093/emboj/18.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westergaard UB, Sorensen ES, Hermey G, Nielsen MS, Nykjaer A, Kirkegaard K, Jacobsen C, Gliemann J, Madsen P, Petersen CM. Functional organization of the sortilin Vps10p domain. J Biol Chem. 2004;279:50221–50229. doi: 10.1074/jbc.M408873200. [DOI] [PubMed] [Google Scholar]
- 6.Tyler-McMahon BM, Boules M, Richelson E. Neurotensin: peptide for the next millennium. Regul Pept. 2000;93:125–136. doi: 10.1016/s0167-0115(00)00183-x. [DOI] [PubMed] [Google Scholar]
- 7.Dicou E, Vincent JP, Mazella J. Neurotensin receptor-3/sortilin mediates neurotensin-induced cytokine/chemokine expression in a murine microglial cell line. J Neurosci Res. 2004;78:92–99. doi: 10.1002/jnr.20231. [DOI] [PubMed] [Google Scholar]
- 8.Martin S, Navarro V, Vincent JP, Mazella J. Neurotensin receptor-1 and −3 complex modulates the cellular signaling of neurotensin in the HT29 cell line. Gastroenterology. 2002;123:1135–1143. doi: 10.1053/gast.2002.36000. [DOI] [PubMed] [Google Scholar]
- 9.Martin S, Vincent JP, Mazella J. Involvement of the neurotensin receptor-3 in the neurotensin-induced migration of human microglia. J Neurosci. 2003;23:1198–1205. doi: 10.1523/JNEUROSCI.23-04-01198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson SK, Christensen S, Raarup MK, Ryan RO, Nielsen MS, Olivecrona G. Endocytosis of apolipoprotein A-V by members of the low density lipoprotein receptor and the VPS10p domain receptor families. J Biol Chem. 2008;283:25920–25927. doi: 10.1074/jbc.M802721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quistgaard EM, Madsen P, Groftehauge MK, Nissen P, Petersen CM, Thirup SS. Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain. Nat Struct Mol Biol. 2009;16:96–98. doi: 10.1038/nsmb.1543. [DOI] [PubMed] [Google Scholar]
- 12.Andersen JL, Schrøder TJ, Christensen S, Strandbygård D, Pallesen LT, García-Alai MM, Lindberg S, Langgård M, Eskildsen JC, David L, Tagmose L, Simonsen KB, Maltas PJ, Rønn LCB, de Jong IEM, Malik IJ, Egebjerg J, Karlsson J-J, Uppalanchi S, Sakumudi DR, Eradi P, Watson SP, Thirup S. Identification of the first small molecule ligand of the neuronal receptor sortilin; and structure determination of the receptor-ligand complex. Acta Cryst. 2014;D70:451–460. doi: 10.1107/S1399004713030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder TJ, Christensen S, Lindberg S, Langgard M, David L, Maltas PJ, Eskildsen J, Jacobsen J, Tagmose L, Simonsen KB, Biilmann Ronn LC, de Jong IE, Malik IJ, Karlsson JJ, Bundgaard C, Egebjerg J, Stavenhagen JB, Strandbygard D, Thirup S, Andersen JL, Uppalanchi S, Pervaram S, Kasturi SP, Eradi P, Sakumudi DR, Watson SP. The identification of AF38469: an orally bioavailable inhibitor of the VPS10P family sorting receptor Sortilin. Bioorg Med Chem Lett. 2014;24:177–180. doi: 10.1016/j.bmcl.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins N, Parekh RB, James DC. Getting the glycosylation right: implications for the biotechnology industry. Nat Biotechnol. 1996;14:975–981. doi: 10.1038/nbt0896-975. [DOI] [PubMed] [Google Scholar]
- 15.Quistgaard EM, Thirup SS. Sequence and structural analysis of the Asp-box motif and Asp-box beta-propellers; a widespread propeller-type characteristic of the Vps10 domain family and several glycoside hydrolase families. BMC Struct Biol. 2009;9:46. doi: 10.1186/1472-6807-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermey G, Sjogaard SS, Petersen CM, Nykjaer A, Gliemann J. Tumour necrosis factor alpha-converting enzyme mediates ectodomain shedding of Vps10p-domain receptor family members. Biochem J. 2006;395:285–293. doi: 10.1042/BJ20051364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazella J, Chabry J, Zsurger N, Vincent JP. Purification of the neurotensin receptor from mouse brain by affinity chromatography. J Biol Chem. 1989;264:5559–5563. [PubMed] [Google Scholar]
- 18.Zsurger N, Mazella J, Vincent JP. Solubilization and purification of a high affinity neurotensin receptor from newborn human brain. Brain Res. 1994;639:245–252. doi: 10.1016/0006-8993(94)91737-x. [DOI] [PubMed] [Google Scholar]
- 19.Eriste E, Norberg A, Nepomuceno D, Kuei C, Kamme F, Tran DT, Strupat K, Jornvall H, Liu C, Lovenberg TW, Sillard R. A novel form of neurotensin post-translationally modified by arginylation. J Biol Chem. 2005;280:35089–35097. doi: 10.1074/jbc.M502567200. [DOI] [PubMed] [Google Scholar]
- 20.Petersen CM, Nielsen MS, Nykjaer A, Jacobsen L, Tommerup N, Rasmussen HH, Roigaard H, Gliemann J, Madsen P, Moestrup SK. Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J Biol Chem. 1997;272:3599–3605. doi: 10.1074/jbc.272.6.3599. [DOI] [PubMed] [Google Scholar]
- 21.W Kabsch. In international tables for crystallography. 2001. Volume F, Chapter 25.2.9., XDS, (ed. Arnold, E.) Kluwer Academic Publishers: Dordrecht. [Google Scholar]
- 22.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Cryst. 2000;D56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 27.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLano WL. The pymol user's manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 29.Sottrup-Jensen L. Determination of halfcystine in proteins as cysteine from reducing hydrolyzates. Biochem Mol Biol Int. 1993;30:789–794. [PubMed] [Google Scholar]
- 30.Bohne-Lang A, von der Lieth CW. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 2005;33:W214–W219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information