Figure 1.
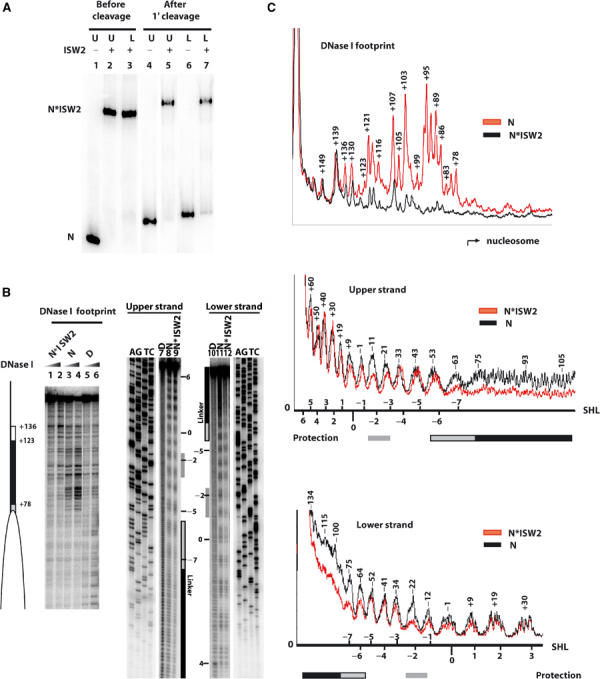
ISW2 binds to three distinct sites on nucleosomes. (A) Sucrose gradient-purified nucleosomes (40 nM) assembled (75-N-0) onto end-labeled 222 bp DNA (upper and lower refer to the opposite strands of DNA) were incubated with 100 nM ISW2. After spin column purification, samples were loaded onto a 4% native polyacrylamide gel before (lanes 1–3) and after (lanes 4–7) cleavage with hydroxyl radicals. Bands correspond to nucleosome alone (N) and nucleosome bound by ISW2 (N*ISW2). (B) End-positioned mononucleosomes reconstituted with either a 250 bp (0-N-103, 90 nM, lanes 1–6) or 222 bp end-labeled DNA (75-N-0, 40 nM, lanes 7–12) were incubated with 150 and 100 nM of ISW2, respectively. Samples were analyzed on a 6.5% polyacrylamide gel containing 8 M urea along with the sequencing ladder of the same DNA, and lanes 1, 2, 9, and 12 all contained ISW2 and nucleosomes. The white ellipse indicates the nucleosome position, and the white and black rectangles indicate the region of ISW2 protection on the linker DNA for DNase I footprinting. For the hydroxyl radical footprinting, the nucleosomal superhelical positions are indicated on the side, and the black (linker DNA) and gray (nucleosomal) boxes indicate regions of ISW2 protection. (C) Quantitation of the DNase I and hydroxyl radical protection patterns of nucleosomes with (N*ISW2) and without (N) ISW2 added are shown. The numbers above the peaks indicate the number of base pairs from the dyad axis. The superhelical location (SHL) of the nucleosome is depicted for the hydroxyl radical footprinting for both upper and lower strands.
