KIFC3, a minus end–directed kinesin motor, recruits the ubiquitin-specific protease USP47, a protease that removes ubiquitin chains from substrates, to epithelial adherens junctions. This process suppresses the ubiquitination and resultant degradation of E-cadherin, thereby maintaining stable cell–cell adhesion in epithelial sheets.
Abstract
The adherens junction (AJ) plays a crucial role in maintaining cell–cell adhesion in epithelial tissues. Previous studies show that KIFC3, a minus end–directed kinesin motor, moves into AJs via microtubules that grow from clusters of CAMSAP3 (also known as Nezha), a protein that binds microtubule minus ends. The function of junction-associated KIFC3, however, remains to be elucidated. Here we find that KIFC3 binds the ubiquitin-specific protease USP47, a protease that removes ubiquitin chains from substrates and hence inhibits proteasome-mediated proteolysis, and recruits it to AJs. Depletion of KIFC3 or USP47 promotes cleavage of E-cadherin at a juxtamembrane region of the cytoplasmic domain, resulting in the production of a 90-kDa fragment and the internalization of E-cadherin. This cleavage depends on the E3 ubiquitin protein ligase Hakai and is inhibited by proteasome inhibitors. E-cadherin ubiquitination consistently increases after depletion of KIFC3 or USP47. These findings suggest that KIFC3 suppresses the ubiquitination and resultant degradation of E-cadherin by recruiting USP47 to AJs, a process that may be involved in maintaining stable cell–cell adhesion in epithelial sheets.
INTRODUCTION
Microtubules interact with cell junctions via their plus ends or minus ends (Stehbens et al., 2006; Ligon and Holzbaur, 2007; Shaw et al., 2007; Meng et al., 2008; Baines et al., 2009; Bellett et al., 2009). Microtubules appear to contribute to molecular trafficking between cell junctions and cytoplasm, a function that is carried out by various kinesin motors or dynein. Even cell adhesion proteins, such as N-cadherin (Mary et al., 2002; Teng et al., 2005) and desmosomal cadherins (Nekrasova et al., 2011), are delivered to the junctions by kinesins. Our previous studies showed that the kinesin family member C3 (KIFC3), a minus end–directed motor (Noda et al., 2001), migrates into the adherens junctions (AJs) of epithelial cells. This migration depends on a recently identified microtubule minus end–binding protein: calmodulin-regulated, spectrin-associated protein 3 (CAMSAP3), which is also called Nezha (Meng et al., 2008; Baines et al., 2009; Tanaka et al., 2012; Nagae et al., 2013). A fraction of CAMSAP3 is localized to AJs through its association with the E-cadherin–p120-catenin–pleckstrin homology domain–containing family A member 7 (PLEKHA7) complex and nucleates microtubule growth from the AJ (Meng et al., 2008). Whether the KIFC3 molecules that move along this population of microtubules carry any cargo to the AJs, however, remains unknown, although this kinesin was found to transport annexin XIIIb to the apical membranes (Noda et al., 2001).
Classical cadherins, including E-cadherin, are key cell adhesion receptors in maintaining the AJ (Takeichi, 2014). For dynamic regulation of AJs, cadherin molecules are dynamically turned over (Delva and Kowalczyk, 2009; Wirtz-Peitz and Zallen, 2009). One of the regulators for cadherin turnover is p120-catenin, which directly binds to a juxtamembrane region of the cadherin cytoplasmic domain (Reynolds, 2007). When p120-catenin is removed, the stability of cadherins in the plasma membrane is greatly reduced, leading to their internalization (Xiao et al., 2007). Ubiquitination of cadherins is another mechanism for stimulating their trafficking (Yang et al., 2006; Schaefer et al., 2012), and several pathways to degrade ubiquitinated cadherins have been identified (Palacios et al., 2005; Janda et al., 2006; Shen et al., 2008).
In the present study, we sought molecules that could be transported by KIFC3 to AJs, and we identified the ubiquitin-specific protease 47 (USP47) as a binding partner for this kinesin. Ubiquitin-specific proteases are a subclass of cysteine proteases that catalyze the removal of ubiquitin from substrates, thus counteracting the activity of E3 ubiquitin ligases. USP47 is one of their subtypes, and it is known to regulate various cell functions (Buus et al., 2009; Peschiaroli et al., 2010; Parsons et al., 2011; Zhang et al., 2013). Our detailed analysis of the function of the KIFC3-USP47 complex suggests that it suppresses E-cadherin degradation and internalization. These findings also lead us to propose a new function of p120-catenin in stabilizing E-cadherin.
RESULTS
KIFC3 recruits USP47 to cell junctions
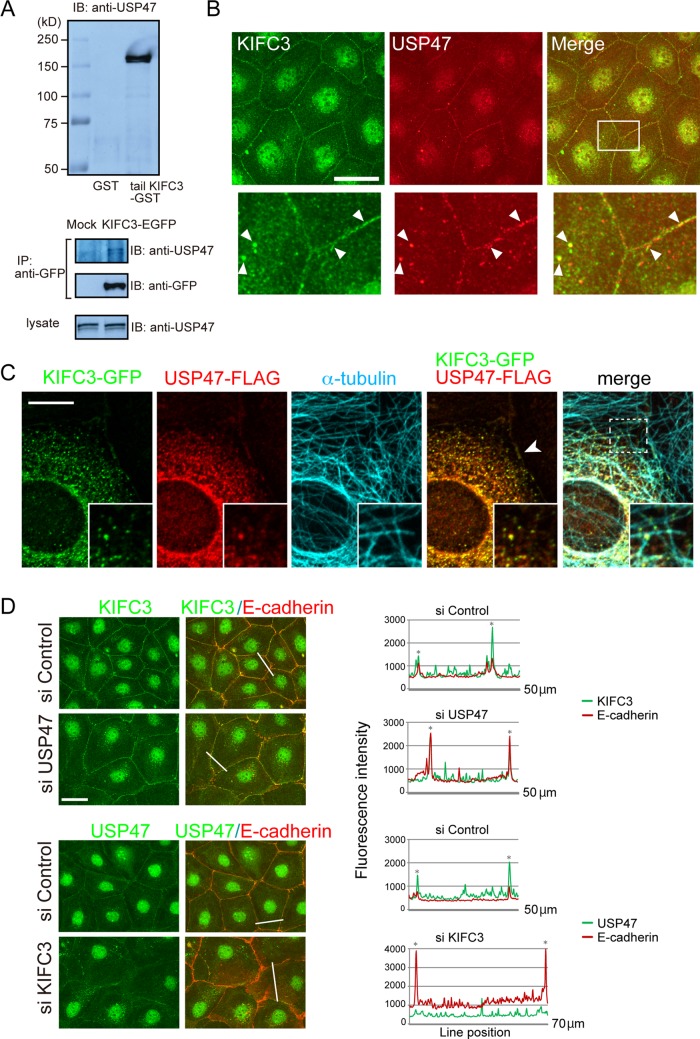
To identify molecules transported by KIFC3, we carried out pull-down assays using glutathione S-transferase (GST) fusion of the KIFC3 tail domain, which is supposed to bind cargo molecules (Hirokawa et al., 2009). Mass spectrographic analysis of the materials precipitated from a lysate of Caco-2 cells showed that the ubiquitin-specific protease USP47 was pulled down with the KIFC3 tail (Supplemental Figure S1A). Western blot analysis using antibodies specific for USP47 confirmed the binding of USP47 to the KIFC3 tail (Figure 1A, top). KIFC3 tagged with enhanced green fluorescent protein (KIFC3-EGFP), which was ectopically expressed in Caco-2 cells, also coprecipitated with endogenous USP47 (Figure 1A, bottom).
FIGURE 1:
KIFC3 binds USP47, and recruits it to cell junctions. (A) Detection of the interaction between of KIFC3 and USP47 by GST pull-down assay (top) and immunoprecipitation (bottom). Top, GST- or tail-KIFC3-GST-immobilized beads were incubated with a Caco-2 cell lysate, and the proteins bound to the beads were analyzed by Western blotting. Bottom, Caco-2 cells were transfected with EGFP or EGFP-KIFC3 expression vector, and their lysates were subjected to immunoprecipitation with anti-GFP antibody. (B) Double immunostaining for endogenous KIFC3 and USP47. Bottom, enlarged images of the boxed area. White arrowheads point to examples of the puncta containing both KIFC3 and USP47. Scale bar, 50 μm. (C) Overlapping distribution of exogenous KIFC3-GFP and USP47-FLAG introduced into Caco-2 cells. Microtubules are also coimmunostained. Insets, enlarged images of the boxed area. Arrowhead indicates junction-associated signals of the two molecules. Scale bars, 10 μm. (D) Distribution of KIFC3 or USP47 after siRNA-mediated depletion of the other. Cells were double immunostained with antibodies specific to E-cadherin and KIFC3 or USP47. Immunostained signals for E-cadherin and KIFC3 or USP47 were scanned along the lines indicated, and data are shown at the right. Asterisks, cell–cell contact points. Scale bars, 50 μm.
Then we examined the subcellular localization of KIFC3 and USP47. Immunostaining for these molecules detected punctate signals both in the cytoplasm and along cell junctions (Figure 1B). Close-up views indicated that KIFC3 and USP47 puncta often overlapped with one another at these sites. These KIFC3- and USP47-immunoreactrive signals faded out when each molecule was depleted with specific small interfering RNAs (siRNAs; Supplemental Figure S1B). To further confirm that the immunostaining signals represent the correct distribution of these molecules, we exogenously expressed KIFC3-GFP and USP47 tagged with FLAG (USP47-FLAG) in Caco-2 cells and found that they coclustered both in the cytoplasm and at cell junctions (Figure 1C). Furthermore, the KIFC3-USP47 coclusters were associated with microtubules (Figure 1C). The junctional localization of KIFC3 was confirmed by double immunostaining for KIFC3-EGFP and E-cadherin (Supplemental Figure S1C).
Double staining for E-cadherin and endogenous KIFC3 or USP47 also showed their overlapping distributions (Figure 1D), indicating that the KIFC3-USP47 complex accumulates at the AJs or zonula adherens (ZA), the epithelial form of an AJ. To determine whether KIFC3 or USP47 is responsible for their junctional localization, we examined the effect of depletion of KIFC3 or USP43 on the distribution of the other. Removal of USP47 had no effect on the localization of KIFC3, whereas KIFC3 loss tended to disturb the junctional accumulation of USP47 (Figure 1D). These observations suggest that USP47 relocates to the AJ via its association with KIFC3, not vice versa.
Depletion of KIFC3 or USP47 destabilizes junctional E-cadherin
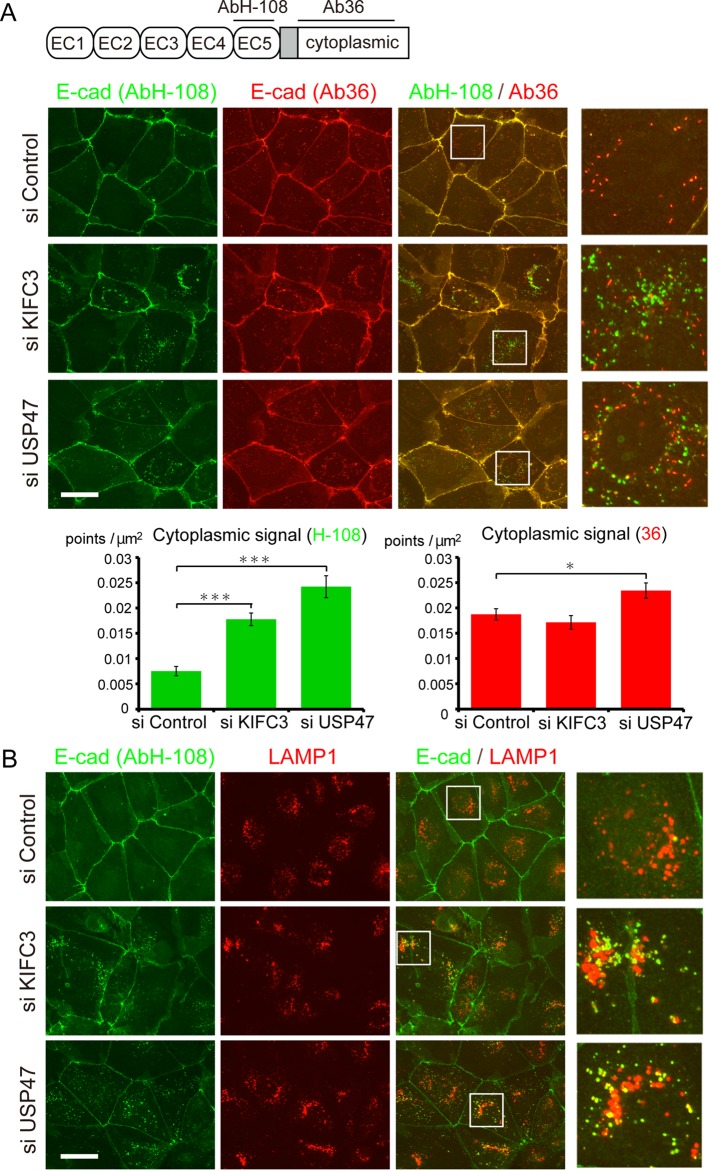
During the foregoing observations, we noticed the tendency that the shape of cell junction delineated with E-cadherin immunosignals became irregular, with a concomitant increase of its cytoplasmic signals after depletion of KIFC3 or USP47. We therefore analyzed these phenomena in more detail, using two kinds of antibodies specific to E-cadherin: polyclonal antibody H-108 (AbH-108), which recognizes amino acids 600–707 of the extracellular domain (Ronzitti et al., 2004; numbered according to the unprocessed human E-cadherin), and a monoclonal antibody called clone 36 (Ab36), specific to the cytoplasmic domain (Kim et al., 2012; Figure 2A and Supplemental Figure S1D). In control cells, although these two antibodies stained the junctional E-cadherin similarly, they reacted differently with intracellular E-cadherin. Ab36 detected cytoplasmic vesicle–like signals, whereas AbH-108 hardly detected such signals (Figure 2A). After depletion of KIFC3 or USP47, however, AbH-108 detected E-cadherin–positive intracellular vesicles, whereas the vesicles detected by Ab36 did not significantly increase (Figure 2A). This suggests that the two antibodies recognize distinct populations of E-cadherin. Actually, only 13% (n = 8) of the vesicles reacted with both antibodies. It is likely that Ab36-positive intracellular signals represent E-cadherin populations undergoing ordinary turnover, as they occur in control cells, whereas AbH-108 detects acutely internalized E-cadherin. In fact, AbH-108–positive vesicles did not colocalize with early endosomal markers such as EEA1 (unpublished data). To further characterize the AbH-108–positive vesicles, we doubly immunostained cells for E-cadherin and LAMP1, a lysosomal protein, and found that ∼50% (n = 8) of AbH-108 signals overlapped with LAMP1 signals (Figure 2B). This suggests that AbH-108–positive E-cadherin molecules tend to be trapped in lysosomes.
FIGURE 2:
Internalization of E-cadherin after KIFC3 or USP47 depletion. (A) Top, sites recognized by the antibodies AbH-108 and Ab36 at the extracellular and intracellular (cytoplasmic) regions of human E-cadherin, respectively, drawn schematically. EC1–EC5 represent the subdomains of the extracellular domain. Middle, cells treated with control siRNA or KIFC3- or USP47-specific siRNAs were double immunostained with AbH-108 and Ab36. Right, enlarged images of the boxed areas. Graphs show quantitation of E-cadherin vesicles in control and KIFC3- or USP47-depleted cells (n = 30). Values indicate mean ± SEM. ***p < 0.0005, *p < 0.05. (B) Cells treated as in A were double immunostained for E-cadherin, using AbH-108, and LAMP1. Right, enlarged images of the boxed areas. Scale bars, 50 μm.
To confirm the specificity of siRNA targeting in these observations, we carried out rescue experiments. Expression of murine KIFC3-FLAG in cells treated with KIFC3 siRNA or an siRNA-resistant mutant of USP47-FLAG in cells treated with USP47 siRNA suppressed the generation of AbH-108–positive E-cadherin vesicles (Supplemental Figure S2).
KIFC3/USP47 depletion enhances formation of a unique E-cadherin fragment
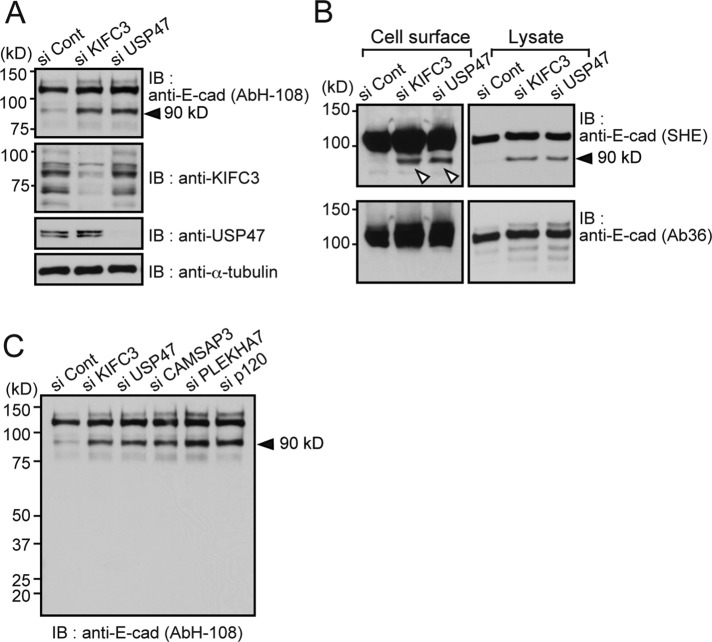
We investigated whether the E-cadherin internalization observed in KIFC3- and USP47-depleted cells was accompanied by any specific form of degradation. Western blot analysis using AbH-108 showed that a band migrating to around the 90-kDa position increased after KIFC3 or USP47 depletion (Figure 3A). Similar results were reproduced using multiple siRNAs (Supplemental Figure S3A), as well as the monoclonal antibodies SHE78-7 and HECD1, which recognize the EC1 and EC2 domains of the E-cadherin extracellular region, respectively (Shiraishi et al., 2005; Supplemental Figures S1D and S3B, top). On the other hand, neither Ab36 nor the antibody 24E10, which recognizes the cytoplasmic sequences surrounding amino acid 780 (Supplemental Figure S1D; Ye et al., 2013), detected the 90-kDa band, although they did detect native E-cadherin on the same blots (Supplemental Figure S3B, bottom). These observations suggest that the 90-kDa fragment of E-cadherin was produced by cleavage at a site between the transmembrane domain and the 24E10 epitope (Supplemental Figure S1D). To determine whether the 90-kDa fragment is still localized in the plasma membrane, we labeled surface proteins on live Caco-2 cells by biotinylation and found that not only the native E-cadherin, but also the 90-kDa fragment was biotinylated (Figure 3B), suggesting that cleavage occurs when the molecules stay in the plasma membrane. Ab36, again, did not detect the 90-kDa band from biotinylated samples.
FIGURE 3:
Generating a 90-kDa E-cadherin fragment by KIFC3 or USP47 depletion. (A) Western blot detection of E-cadherin using AbH-108 after KIFC3 or USP47 depletion. Levels of KIFC3 or USP47 in respective samples are also shown. α-Tubulin was used as a loading control. Black arrowheads point to the 90-kDa E-cadherin fragment. (B) Detection of 90-kDa fragments from cell surface protein–biotinylated samples. Left, isolated biotinylated proteins were subjected to Western blotting, from which E-cadherin was detected using antibodies recognizing its extracellular (SHE78-7 [SHE]) or intracellular (Ab36) region. Only SHE78-7 detects the 90-kDa band (white triangles). Right, total cell lysates. Lower kilodalton bands, detected by Ab36, do not correspond to the 90-kDa band, as assessed by close comparisons of their positions. (C) Effects of CAMSAP3, PLEKHA7, and p120-catenin knockdown on production of the 90-kDa fragment.
Localization of KIFC3 to AJs depends on the cadherin–p120-catenin–PLEKHA7–CAMSAP3 complex (Meng et al., 2008). We therefore examined the effects of siRNA-mediated knockdown of PLEKHA7 and p120-catenin, as well as of CAMSAP3. Depletion of all of these proteins enhanced production of the 90-kDa band (Figure 3C and Supplemental Figure S3B), as well as E-cadherin internalization, which can be detected by immunostaining with AbH-108 (unpublished data). These results are consistent with the idea that the entire machinery to promote the KIFC3 localization to AJs is involved in suppressing production of the 90-kDa fragment.
Proteasomes mediate the initial E-cadherin fragmentation
To identify proteolytic mechanisms that could produce the 90-kDa fragment, we examined the effects of various protease inhibitors on this process using KIFC3-depleted cells. Presenilin/γ-secretase and matrix metalloprotease are believed to cleave E-cadherin at the C- and N-terminal sides, respectively, of the transmembrane domain (Marambaud et al., 2002). However, the γ-secretase inhibitor L685,458 and the matrix metalloprotease inhibitor GM6001 had no effect on the production of 90-kDa fragments (Supplemental Figure S3C, left). Caspase-3 and calpain are known to cleave the linkage between amino acids 750 (D) and 751 (N; Steinhusen et al., 2001) and between 781 (E) and 782 (V; Ye et al., 2013), respectively, at the E-cadherin cytoplasmic domain. However, the caspase-3 inhibitor Z-DEVD-FMK and the calpain inhibitors MDL and calpetin had little effect on the 90-kDa fragmentation (Supplemental Figure S3C, right).
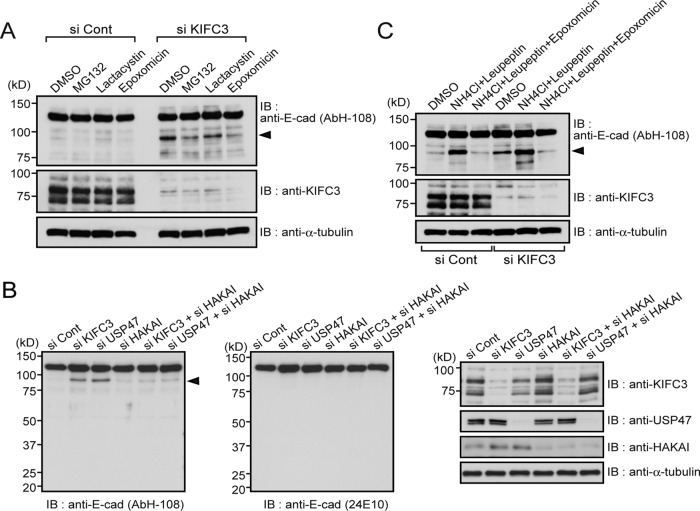
Then we tested the effects of three proteasome inhibitors—MG-132, lactacystin, and epoxomicin. All of these reagents tended to suppress the generation of the 90-kDa fragment (Figure 4A), although lactacystin showed a relatively weaker effect than the others. These results suggest that the proteasome is involved in the cleavage of E-cadherin to produce its 90-kDa fragment. The proteasome targets ubiquitin-tagged proteins for degradation. Because E-cadherin is known to be ubiquitinated by Hakai, an E3 ubiquitin ligase (Fujita et al., 2002), we examined whether Hakai is involved in the KIFC3/USP47 depletion–dependent formation of 90-kDa fragments. Hakai depletion using its specific siRNAs clearly suppressed this process (Figure 4B), consistent with the idea that the proteasome mediates production of the 90-kDa fragment. Involvement of proteasomes in stabilization of E-cadherin–mediated junctions has also been suggested by other studies (Tsukamoto and Nigam, 1999; Davis et al., 2003; Saitoh et al., 2009).
FIGURE 4:
Production of the 90-kDa E-cadherin fragment depends on proteasome and Hakai. (A) Effects of proteasome inhibitors on 90-kDa fragment production. Caco-2 cells were transfected with control or KIFC3-specific siRNA. At 7 h after transfection, cells were passaged. After 24 h, dimethyl sulfoxide, 1 μM MG132, 10 μM lactacystin, or 100 nM epoxomycin was added to culture medium. At 16 h later, cells were harvested and subjected to Western blotting assay using the antibodies indicated. (B) Effects of Hakai depletion on 90-kDa fragment production. At 48 h after transfection of cells with indicated siRNAs, cells were harvested and subjected to Western blotting assay using the antibodies indicated. Knockdown efficiency for each molecule is shown on the far right. (C) Effects of proteasome and lysosome inhibitors on 90-kDa fragment production. Cells were treated with siRNAs and subsequently with 10 mM NH4Cl and 21 μM leupeptin with or without 100 nM epoxomycin, according to the same protocol as in A.
However, this putative mechanism appears to conflict with the general consensus that the proteasome works for degradation of intracellular soluble proteins (Glickman and Ciechanover, 2002; Tanno and Komada, 2013) and with the observation that internalized cadherin molecules are detected in the lysosome, the organelle that is known to degrade transmembrane proteins (Hicke and Dunn, 2003). To solve the inconsistency among these observations, we looked at the effect of the lysosomal inhibitor leupeptin on production of the 90-kDa fragment. Surprisingly, the treatment of cells with leupeptin resulted in a dramatic increase in the 90-kDa band not only in control cells, but also in KIFC3-depleted cells (Figure 4C), implying that, in normal situations, the 90-kDa fragments are further degraded by lysosomes. The proteasome inhibitor epoxomicin suppressed this process, suggesting that the proteasome-mediated cleavage of E-cadherin is prerequisite for its lysosomal degradation. Thus there seems to be a two-step degradation of E-cadherin: initially, a partial cleavage of E-cadherin by proteasomes that produces the 90-kDa fragment, and then further degradation of this fragment by lysosomes. These observations also confirmed that formation of the 90-kDa fragment takes place even in the presence of KIFC3 to a certain degree.
Enhancement of E-cadherin ubiquitination by KIFC3/USP47 depletion
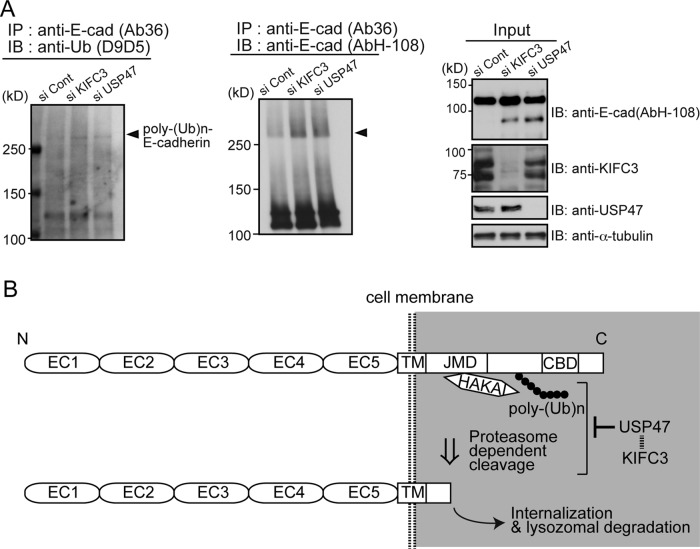
Finally, we tested whether KIFC3/USP47 indeed regulates E-cadherin ubiquitination. Western blots of immunoprecipitated E-cadherin, probed with antibodies specific to the K48-linked ubiquitin chain that serves to direct the target proteins for proteasome-dependent degradation (Tanno and Komada, 2013), detected a band at a region of high molecular mass, and the band intensity increased in KIFC3/USP47-depleted samples (Figure 5A). Moreover, smear signals, a hallmark of ubiquitinated proteins, increased in E-cadherin blots at positions above the K48-linked ubiquitin-positive band. These observations confirmed the importance of the KIFC3/USP47 system in suppressing E-cadherin ubiquitination.
FIGURE 5:
KIFC3 and USP47 are required to suppress E-cadherin ubiquitination. (A) Western blotting detection of ubiquitin from E-cadherin using D9D5, an antibody that detects only the polyubiquitin chains formed by Lys-48 residue linkage (left), and that of putative ubiquitinated E-cadherin (middle). Left, cells were harvested at 48 h after transfection with siRNAs. From their lysates, E-cadherin was immunoprecipitated using Ab36, and the precipitates were analyzed to detect polyubiquitinated bands. Middle, E-cadherin was immunoprecipitated using Ab36, and the precipitates were analyzed with AbH-108. Smeared E-cadherin bands with high molecular sizes increase after KIFC3 or USP47 depletion. Arrowheads indicate the position of D9D5-positive bands. Right, input samples. (B) Working model for the role of KIFC3 and USP47 in E-cadherin homeostasis. E-cadherin is polyubiquitinated by Hakai and cleaved at an intracellular site by proteasomes. USP47 inhibits these processes to maintain E-cadherin at cell–cell contacts. The ubiquitinated site is drawn hypothetically.
DISCUSSION
On the basis of the present observations, we propose that the following molecular events regulate E-cadherin stability. USP47 is transported to AJs by KIFC3, and it counteracts the action of E3 ubiquitin ligases, including Hakai, in order to suppress E-cadherin ubiquitination (Figure 5B). This system is, however, canceled by loss of KIFC3 or USP47, resulting in enhanced ubiquitination of E-cadherin. The ubiquitinated E-cadherin is targeted by proteasomes to produce a 90-kDa fragment, and the fragment is further degraded by the lysosome. In this model, however, the following points remain open: how E-cadherin as a transmembrane protein is targeted by proteasomes, and why E-cadherin is only partially cleaved upon proteasome-dependent digestion. We suspect that proteasomes do recognize a ubiquitinated E-cadherin as a substrate but cannot incorporate the entire E-cadherin molecule into their core particle for digestion because E-cadherin is trapped in the plasma membrane. This might have resulted in the cleavage of E-cadherin only at its cytoplasmic tail. The previous finding that an isolated E-cadherin juxtamembrane domain (JMD) is ubiquitinated and then degraded with proteasomes (Hartsock and Nelson, 2012) is consistent with our finding that the cleavage that produced the 90-kDa fragment occurred within the JMD.
Intriguingly, the increase of the 90-kDa fragments was not accompanied by a reduction of the full-length E-cadherin in Western blotting. Similarly, immunostaining intensity for E-cadherin was not necessarily decreased at cell–cell contacts of USP47- or KIFC3-depleted cells, although the junctional morphology tended to be perturbed. The observations suggest that cells may promptly supply native E-cadherin molecules to the cell membranes in compensation for their loss, but such rapid replacement of E-cadherin might have caused destabilization of the junctional structures.
p120-catenin plays a critical role in preventing cadherins from internalization (Reynolds, 2007; Xiao et al., 2007). It has been shown that p120-catenin stabilizes cadherins by masking endocytic signals embedded in the JMD (Miyashita and Ozawa, 2007; Nanes et al., 2012). Our present results suggest an additional mechanism of p120-catenin–dependent E-cadherin stabilization. Not only KIFC3/USP47, but also p120-catenin and PLEKHA7 were required to protect E-cadherin against its degradation into the 90-kDa fragment, and the p120-catenin–PLEKHA7 complex played a role in recruiting CAMSAP3 to AJs (Meng and Takeichi, 2009). p120-catenin may serve E-cadherin stabilization by leading the KIFC3–USP47 complex to these junctions. Of note, the KIFC3–USP47 complex was detected throughout the cytoplasm. This implies that the targets of this complex may not be restricted to E-cadherin. In fact, USP47 appears to regulate multiple cellular processes (Buus et al., 2009; Peschiaroli et al., 2010; Parsons et al., 2011; Zhang et al., 2013). KIFC3 could be broadly used for USP47 trafficking.
MATERIALS AND METHODS
Cell culture
Caco-2 cells were maintained in DMEM/Ham's F-12 (Wako Pure Chemical, Osaka, Japan) containing 10% fetal bovine serum.
Plasmids
The expression vector for pEGFP-C1-mouse KIFC3 (mKIFC3) was a kind gift from N. Hirokawa (University of Tokyo, Tokyo, Japan). The N-terminal 510 base pairs of mKIFC3 (tail-KIFC3) was subcloned into a modified pGEX-4T vector (GE Healthcare, Little Chalfont, United Kingdom). Glutathione S-transferase (GST) was fused to the C-terminus of tail-KIFC3. Full-length mKIFC3 was subcloned into a pCANw-FLAG vector. FLAG was fused to the C-terminus of full-length mKIFC3. Human KIFC3 (hKIFC3) was newly cloned from a Caco-2 cDNA library, which was prepared using RNeasy Mini Kit (Qiagen, Venlo, Netherlands) and SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, CA). hKIFC3 was subcloned into a pEGFP-C vector (Clontech Laboratories, Mountain View, CA) or pTagGFP2-N vector (Evrogen, Moscow, Russia). Human USP47, inserted into pcDNA3.1-FLAG, was a kind gift from G. Melino (University of Rome, Rome, Italy; Peschiaroli et al., 2010). To prepare a siRNA-resistant mutant of USP47 cDNA, we amplified a mutant clone in which the 408th codon in the siRNA#3-target sequences was changed from TCT to AGC by the inverse PCR method using a KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan). The mutations did not change the amino acid sequence of the resulting protein. Transfection of cells was performed using FuGENE HD Transfection Reagent (Promega, Madison, WI), FuGENE 6 Transfection Reagent (Promega), or X-tremeGENE HP (Roche Diagnostics, Mannheim, Germany).
Reagents
Epoxomicin was purchased from Sigma-Aldrich (St. Louis, MO). InSolution γ-Secretase Inhibitor X (L-685,458; 1S-Benzyl-4R-[1-(1S-carbamoyl-2-phenethylcarbamoyl)-1S-3-methylbutylcarbamoyl]-2R-hydroxy-5-phenylpentyl}carbamic acid tert-butyl ester), InSolution GM6001, caspase-3 inhibitor I (DEVD-CHO), and MG-132 were purchased from Merck Millipore (Billerica, MA). Clasto-lactacystine β-lactone was purchased from Research & Diagnostics Systems (Minneapolis, MN). Leupeptin was purchased from Peptide Institute (Osaka, Japan). MDL 28170 was purchased from Enzo Life Sciences (Farmingdale, NY). Calpeptin was purchased from Santa Cruz Biotechnology (Dallas, TX). All other chemicals were from Nacalai Tesque (Kyoto, Japan), unless otherwise indicated.
Purification of GST and GST-fused tail-KIFC3
pGEX-4T mock vector and modified pGEX-4T-tail-KIFC3 vector were introduced into DH-5α– competent cells. The cells were spread on ampicillin-containing lysogeny broth plates and incubated at 37ºC overnight. Each colony was inoculated in an ampicillin-containing LB medium and shaken at 37ºC overnight. A small amount of culture medium was added to a new ampicillin-containing LB medium and shaken for 3.5 h at 37ºC (OD600 = 0.9). Isopropyl thiogalactoside, 400 μM, was added to each culture medium and shaken for 3 h at 30ºC. Culture medium was then centrifuged at 8000 × g for 5 min at 4ºC. Each pellet was vortexed with cold phosphate-buffered saline (PBS) containing a protease inhibitor cocktail (Complete EDTA-free, Roche Diagnostics), sonicated for 10 s, and incubated on ice for 30 s. The sonication process was repeated six times. Each cell solution was rotated with 1% Triton X-100 for 30 min at 4ºC and centrifuged at 12,000 × g for 10 min at 4ºC. The supernatant was rotated with Glutathione Sepharose 4B (GE Healthcare) for 1.5 h at 4ºC and centrifuged at 50 × g for 5 min at 4ºC. Beads were washed three times with cold PBS containing the protease inhibitor cocktail and suspended in it.
GST pull-down assay
All processes were performed at 4ºC. Caco-2 cells were washed two times with cold PBS and harvested with cold PBS containing a protease inhibitor cocktail. After centrifugation at 190 × g for 5 min, the pellet was suspended with TNE buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 5% glycerol, 0.5 mM dithiothreitol) containing a protease inhibitor cocktail and transferred to a tube. After rotation for 30 min and centrifugation at 17,000 × g for 5 min, the supernatant was decanted to another tube and centrifuged similarly. After the secondary decantation to a new tube, the supernatant was precleared with GST-immobilized beads by rotation for 1 h. Thereafter the supernatant was prepared by centrifugation at 300 × g for 5 min and mixed with GST-immobilized beads or GST-tail-KIFC3–immobilized beads. Each mixture was rotated for 1.5 h and centrifuged at 100 × g for 5 min. The beads were washed three times with wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 5% glycerol) containing the protease inhibitor cocktail and boiled with 2× SDS buffer (100 mM Tris-HCl, pH 6.8, 2% SDS, 10% β-mercaptoethanol [β-ME], 20% glycerol, and 0.003% bromophenol blue).
Mass spectrometry
Proteins were separated on SDS–PAGE. The gel was stained with 2D-Silver Stain II (Cosmo Bio, Tokyo, Japan), according to the manufacturer's protocol. The gel pieces containing specific bands were cut out and subjected to mass spectrometry analysis.
Preparation and transfection of siRNAs
siRNAs specific to hKIFC3 (#1; SASI_Hs01_00046906, #2; SASI_Hs01_00046907, #3; SASI_Hs01_00046908), human USP47 (#1; SASI_Hs01_00112225, #2; SASI_Hs01_00112227, #3; SASI_Hs02_00350956), human CBLL1/HAKAI (SASI_Hs01_00020900), human CAMSAP3 (SASI_Hs02_00314756), human PLEKHA7 (SASI_Hs01_00054762), and human CTNND2/p120 (SASI_Hs01_00147132) were purchased from Sigma-Aldrich. MISSION siRNA Universal Negative Control (Sigma-Aldrich) was used as a control. siRNA transfection was performed with Lipofectamine RNAiMAX Transfection Reagent (Life Technologies), according to the manufacturer's protocol.
Western blot analysis
Cells were harvested with RIPA buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 25 mM NaF, 1 mM Na3VO4) containing the protease inhibitor cocktail. After centrifugation at 17,000 × g for 20 min at 4ºC, the supernatant was decanted to another tube. Concentration of proteins was measured using a DC Protein Assay (Bio-Rad, Hercules, CA). The supernatant was mixed with 4× SDS buffer (200 mM Tris-HCl, pH 6.8, 4% SDS, 20% β-ME, 40% glycerol, 0.006% BPB) and boiled for 5 min. Proteins were separated on SDS–PAGE and electrotransferred onto Immobilon-P Transfer Membranes (Merck Millipore) using a semidry transferring machine. Membranes were incubated in a blocking buffer (Tris-buffered saline containing 5% skim milk and 0.05% Tween 20) for 1 h at room temperature and blotted with primary antibodies in the blocking buffer at 4ºC overnight. After blotting, the membranes were washed three times with Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and incubated with an anti-mouse or anti-rabbit horseradish peroxidase (HRP)–linked secondary antibody (GE Healthcare) in the blocking buffer for 1 h at room temperature. The membranes were washed four times with TBS-T and contacted Western Lightning Plus-ECL (PerkinElmer, Waltham, MA) or Chemi-Lumi One Super (Nacalai Tesque) for 1 min. Protein bands reacting with antibodies were detected with x-ray film (RX; Fujifilm, Tokyo, Japan).
Immunoprecipitation assay
All processes were performed at 4ºC. To test the binding between KIFC3 and USP47, we transfected Caco-2 cells with pEGFP-C1-mKIFC3. After 48 h, cells were harvested with TNE buffer containing the protease inhibitor cocktail. The solution was centrifuged at 200,000 × g for 30 min, and the supernatant was rotated with Protein G Sepharose (GE Healthcare) for 1 h. Concurrently, anti-GFP antibody (598) in TNE buffer was rotated with Protein G Sepharose. The cell solution was centrifuged at 1900 × g for 1 s. The supernatant was transferred to another tube and rotated with anti-GFP antibody-immobilized Protein G Sepharose for 3 h. After centrifugation at 100 × g for 5 min, the beads were washed three times with cold PBS containing the protease inhibitor cocktail and boiled with 2× SDS buffer.
Detection of ubiquitination
Immunoprecipitation of ubiquitinated proteins was performed based on Swaminathan and Cartwright (2012). At 48 h after transfection of cells with respective siRNAs, the cells were harvested with a lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1% Triton X-100, 0.25% N-ethylmaleimide, 250 mM NaF, 2.5 mM Na3VO4) containing 1% SDS and protease inhibitor cocktail. After centrifugation at 17,000 × g for 20 min at 4ºC, the supernatant was diluted with a minimal amount of lysis buffer. After centrifugation at 17,000 × g for 10 min at 4ºC, the supernatant was subjected to protein concentration measurement. Cell solutions containing 950 μg of proteins were transferred to another tube, added with the lysis buffer up to 5 ml in total, and rotated with anti–E-cadherin antibody–immobilized Protein G-Sepharose for 1 h at 4ºC. After centrifugation at 800 × g for 1 s, the beads were washed sequentially with four different buffers—a lysis buffer containing 0.5 M NaCl, a lysis buffer containing 0.1% SDS, the original lysis buffer, and TBS—before boiling with 2× SDS buffer. Proteins were separated on SDS–PAGE in a gradient gel (SuperSep 3–10%; Wako Pure Chemical) and electrotransferred onto Immun-Blot PVDF Membrane (Bio-Rad Laboratories) using a wet transferring machine at 4ºC overnight. Membranes were incubated in a blocking buffer (TBS-T containing 5% bovine serum albumin) for 1 h at room temperature and blotted with anti-ubiquitin antibody (D9D5) in the blocking buffer at 4ºC overnight. After blotting, the membranes were washed three times with TBS-T and incubated with anti-rabbit HRP-linked secondary antibody in the same blocking buffer for 1 h at room temperature. The membranes were washed four times with TBS-T, and subjected to protein band detection.
Immunocytochemistry
Cells were cultured on collagen I–coated coverslips (AGC Techno Glass, Shizuoka, Japan), washed with PBS or TBS (PBS/TBS), and fixed with 1% paraformaldehyde in PBS/TBS for 10 min at room temperature. After washing with PBS/TBS, the cells were permeabilized with PBS/TBS containing 0.1% Triton (PBS/TBS-T) for 10 min at room temperature. For detection of α-tubulin and LAMP1, cells were fixed with cold methanol at −20ºC for 5 and 20 min, respectively. After washing with PBS/TBS-T, cells were incubated with a blocking buffer (Antibody Diluent, Dako, Glostrup, Denmark) for 30 min at room temperature and incubated with primary antibodies at 4ºC overnight. The cells were then washed with PBS/TBS-T and incubated with secondary antibodies for 1 h at room temperature. After washing with PBS-T 4 times, the coverslips were mounted onto the slides using Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). Fluorescence images were acquired using a Zeiss AxioPlan 2 microscope (Carl Zeiss, Oberkochen, Germany) through a Plan-NEOFLUAR ×40/1.30 numerical aperture (NA) oil-immersion objective lens (Carl Zeiss) and an ORCA-R2 C10600 digital charge-coupled device camera (Hamamatsu Photonics, Shizuoka, Japan). Confocal images were acquired with a Zeiss LSM710 confocal microscope (Carl Zeiss) through a Plan-Apochromat ×63/1.40 NA oil-immersion objective lens (Carl Zeiss). Projection of Z-stack images (5-μm sections for microtubules; 0.3 μm for cell junctions) were acquired using Zen software (Carl Zeiss).
Antibodies
For primary antibodies, we used anti-KIFC3 antibody (H-300; Santa Cruz Biotechnology), anti-USP47 antibody (4E7; Santa Cruz Biotechnology; ab72143; Abcam, Cambridge, UK), anti-GFP antibody (598, Medical and Biological Laboratories, Nagoya, Japan; GF090R, Nacalai Tesque), anti–E-cadherin antibodies (H-108; Santa Cruz Biotechnology; SHE78-7 and HECD-1; Takara Bio, Shiga, Japan; 36; BD Biosciences, San Jose, CA; 24E10; Cell Signaling Technology, Danvers, MA), anti-CBLL1/HAKAI antibody (Proteintech Group, Chicago, IL), anti-PLEKHA7 antibody (Sigma-Aldrich), anti–p120-catenin antibody (BD Biosciences), anti–α-tubulin antibody (DM1A, Sigma-Aldrich; Abcam), anti-LAMP1 antibody (Santa Cruz Biotechnology), and anti-FLAG antibody (M2, Sigma-Aldrich). Rabbit polyclonal anti-CAMSAP3 antibody was produced previously (Tanaka et al., 2012). For secondary antibodies, we used Alexa 488–conjugated anti-mouse secondary antibody, Alexa 555–conjugated anti-mouse secondary antibody, Alexa 568–conjugated anti-mouse secondary antibody, Alexa 488–conjugated anti-rabbit secondary antibody, Alexa 568–conjugated anti-rabbit secondary antibody, Alexa 647–conjugated anti-rabbit secondary antibody, and Alexa 488–conjugated anti-rat secondary antibody (Life Technologies).
Cell surface biotinylation assay
To biotinylate cell surface proteins, Pierce Cell Surface Protein Isolation Kit (Thermo Fisher Scientific, Waltham, MA) was used, according to the manufacturer's protocol.
Quantification of immunostained images
E-cadherin–positive vesicles were automatically detected using ImageJ software (National Institutes of Health, Bethesda, MD) with a home-made plug-in. We used the multiscale variance stabilization transform (MSVST) method together with the isotropic undecimated wavelet transform (IUWT) previously described (Starck et al., 2009). Briefly, vesicle images were transformed three times using IUWT, and the wavelet coefficients were obtained from the transformed images. Denoising was performed by ignoring the coefficients smaller than a manually determined threshold. Denoised vesicle images were reconstructed with the coefficients, and in the reconstructed images, signals stronger than that of the manually determined threshold were taken to be E-cadherin–containing vesicles. To measure immunostaining intensities across cell junctions, we used the plot profile function of Fiji software. To estimate the overlap between E-cadherin vesicles immunostained with different antibodies, we analyzed images with Fiji. The images from two channels were first converted into binary images, respectively. The binary image was multiplied with another original image to show only merged signals. Then merged vesicles were counted.
Statistical analysis
For analysis of the differences between control cells and KIFC3 or USP47 knockdown cells in immunostained images, statistical comparisons were performed using the unpaired Student's t test. Data were considered statistically significant with values of p < 0.05 compared with control.
Supplementary Material
Acknowledgments
We thank N. Hirokawa for KIFC3 cDNA, G. Melino for USP47 cDNA, K. Shinmyozu (Center for Developmental Biology Mass Spectrometry Analysis Unit) for mass spectrometric analysis, Y. Fujita for discussion, and K. Nagata for critical reading of the manuscript. We are also grateful to M. Nomura-Harata, H. Saitou, Y. Inoue, and H. Sylvain for their technical support, as well as to other members of our laboratory for discussion. This work was supported by Grant-in-Aid for Specially Promoted Research 20002009 and Grant-in-Aid for Scientific Research (S) 2522110 from the Japan Society for Promotion of Science to M.T.
Abbreviations used:
- AJ
adherens junction
- CAMSAP3
calmodulin-regulated, spectrin-associated protein3
- KIFC3
kinesin family member C3
- PLEKHA7
pleckstrin homology domain–containing family A member 7
- USP47
ubiquitin-specific peptidase 47.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1245) on September 24, 2014.
REFERENCES
- Baines AJ, Bignone PA, King MD, Maggs AM, Bennett PM, Pinder JC, Phillips GW. The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol Biol Evol. 2009;26:2005–2014. doi: 10.1093/molbev/msp115. [DOI] [PubMed] [Google Scholar]
- Bellett G, Carter JM, Keynton J, Goldspink D, James C, Moss DK, Mogensen MM. Microtubule plus-end and minus-end capture at adherens junctions is involved in the assembly of apico-basal arrays in polarised epithelial cells. Cell Motil Cytoskeleton. 2009;66:893–908. doi: 10.1002/cm.20393. [DOI] [PubMed] [Google Scholar]
- Buus R, Faronato M, Hammond DE, Urbe S, Clague MJ. Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr Biol. 2009;19:1463–1466. doi: 10.1016/j.cub.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E, Kowalczyk AP. Regulation of cadherin trafficking. Traffic. 2009;10:259–267. doi: 10.1111/j.1600-0854.2008.00862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. Competitive regulation of E-cadherin juxtamembrane domain degradation by p120-catenin binding and Hakai-mediated ubiquitination. PLoS One. 2012;7:e37476. doi: 10.1371/journal.pone.0037476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev. Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Janda E, Nevolo M, Lehmann K, Downward J, Beug H, Grieco M. Raf plus TGFbeta-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene. 2006;25:7117–7130. doi: 10.1038/sj.onc.1209701. [DOI] [PubMed] [Google Scholar]
- Kim H, He Y, Yang I, Zeng Y, Kim Y, Seo YW, Murnane MJ, Jung C, Lee JH, Min JJ, et al. delta-Catenin promotes E-cadherin processing and activates beta-catenin-mediated signaling: implications on human prostate cancer progression. Biochim Biophys Acta. 2012;1822:509–521. doi: 10.1016/j.bbadis.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Holzbaur EL. Microtubules tethered at epithelial cell junctions by dynein facilitate efficient junction assembly. Traffic. 2007;8:808–819. doi: 10.1111/j.1600-0854.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary S, Charrasse S, Meriane M, Comunale F, Travo P, Blangy A, Gauthier-Rouviere C. Biogenesis of N-cadherin-dependent cell-cell contacts in living fibroblasts is a microtubule-dependent kinesin-driven mechanism. Mol Biol Cell. 2002;13:285–301. doi: 10.1091/mbc.01-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Ozawa M. Increased internalization of p120-uncoupled E-cadherin and a requirement for a dileucine motif in the cytoplasmic domain for endocytosis of the protein. J Biol Chem. 2007;282:11540–11548. doi: 10.1074/jbc.M608351200. [DOI] [PubMed] [Google Scholar]
- Nagae S, Meng W, Takeichi M. Non-centrosomal microtubules regulate F-actin organization through the suppression of GEF-H1 activity. Genes Cells. 2013;18:387–396. doi: 10.1111/gtc.12044. [DOI] [PubMed] [Google Scholar]
- Nanes BA, Chiasson-MacKenzie C, Lowery AM, Ishiyama N, Faundez V, Ikura M, Vincent PA, Kowalczyk AP. p120-catenin binding masks an endocytic signal conserved in classical cadherins. J Cell Biol. 2012;199:365–380. doi: 10.1083/jcb.201205029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova OE, Amargo EV, Smith WO, Chen J, Kreitzer GE, Green KJ. Desmosomal cadherins utilize distinct kinesins for assembly into desmosomes. J Cell Biol. 2011;195:1185–1203. doi: 10.1083/jcb.201106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda Y, Okada Y, Saito N, Setou M, Xu Y, Zhang Z, Hirokawa N. KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J Cell Biol. 2001;155:77–88. doi: 10.1083/jcb.200108042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F, Tushir JS, Fujita Y, D'Souza-Schorey C. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol. 2005;25:389–402. doi: 10.1128/MCB.25.1.389-402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol Cell. 2011;41:609–615. doi: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Skaar JR, Pagano M, Melino G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene. 2010;29:1384–1393. doi: 10.1038/onc.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB. p120-catenin: past and present. Biochim Biophys Acta. 2007;1773:2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti G, Callegari F, Malaguti C, Rossini GP. Selective disruption of the E-cadherin-catenin system by an algal toxin. Br J Cancer. 2004;90:1100–1107. doi: 10.1038/sj.bjc.6601640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Shirakihara T, Miyazono K. Regulation of the stability of cell surface E-cadherin by the proteasome. Biochem Biophys Res Commun. 2009;381:560–565. doi: 10.1016/j.bbrc.2009.02.098. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Nethe M, Hordijk PL. Ubiquitin links to cytoskeletal dynamics, cell adhesion and migration. Biochem J. 2012;442:13–25. doi: 10.1042/BJ20111815. [DOI] [PubMed] [Google Scholar]
- Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hirsch DS, Sasiela CA, Wu WJ. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J Biol Chem. 2008;283:5127–5137. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- Shiraishi K, Tsuzaka K, Yoshimoto K, Kumazawa C, Nozaki K, Abe T, Tsubota K, Takeuchi T. Critical role of the fifth domain of E-cadherin for heterophilic adhesion with alpha E beta 7, but not for homophilic adhesion. J Immunol. 2005;175:1014–1021. doi: 10.4049/jimmunol.175.2.1014. [DOI] [PubMed] [Google Scholar]
- Starck J-L, Fadili JM, Digel S, Zhang B, Chiang J. Source detection using a 3D sparse representation: application to the Fermi gamma-ray space telescope. Astron Astrophys. 2009;504:641–652. [Google Scholar]
- Stehbens SJ, Paterson AD, Crampton MS, Shewan AM, Ferguson C, Akhmanova A, Parton RG, Yap AS. Dynamic microtubules regulate the local concentration of E-cadherin at cell-cell contacts. J Cell Sci. 2006;119:1801–1811. doi: 10.1242/jcs.02903. [DOI] [PubMed] [Google Scholar]
- Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- Swaminathan G, Cartwright CA. Rack1 promotes epithelial cell-cell adhesion by regulating E-cadherin endocytosis. Oncogene. 2012;31:376–389. doi: 10.1038/onc.2011.242. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev. Mol Cell Biol. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Meng W, Nagae S, Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci USA. 2012;109:20029–20034. doi: 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno H, Komada M. The ubiquitin code and its decoding machinery in the endocytic pathway. J Biochem. 2013;153:497–504. doi: 10.1093/jb/mvt028. [DOI] [PubMed] [Google Scholar]
- Teng J, Rai T, Tanaka Y, Takei Y, Nakata T, Hirasawa M, Kulkarni AB, Hirokawa N. The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat Cell Biol. 2005;7:474–482. doi: 10.1038/ncb1249. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Nigam SK. Cell-cell dissociation upon epithelial cell scattering requires a step mediated by the proteasome. J Biol Chem. 1999;274:24579–24584. doi: 10.1074/jbc.274.35.24579. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Zallen JA. Junctional trafficking and epithelial morphogenesis. Curr Opin Genet Dev. 2009;19:350–356. doi: 10.1016/j.gde.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Oas RG, Chiasson CM, Kowalczyk AP. Role of p120-catenin in cadherin trafficking. Biochim Biophys Acta. 2007;1773:8–16. doi: 10.1016/j.bbamcr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, Broglio K, Berry DA, Hung MC. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26:7269–7282. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Tian H, Lange AR, Yearsley K, Robertson FM, Barsky SH. The genesis and unique properties of the lymphovascular tumor embolus are because of calpain-regulated proteolysis of E-cadherin. Oncogene. 2013;32:1702–1713. doi: 10.1038/onc.2012.180. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev 27, 1581–1595. 2013 doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.