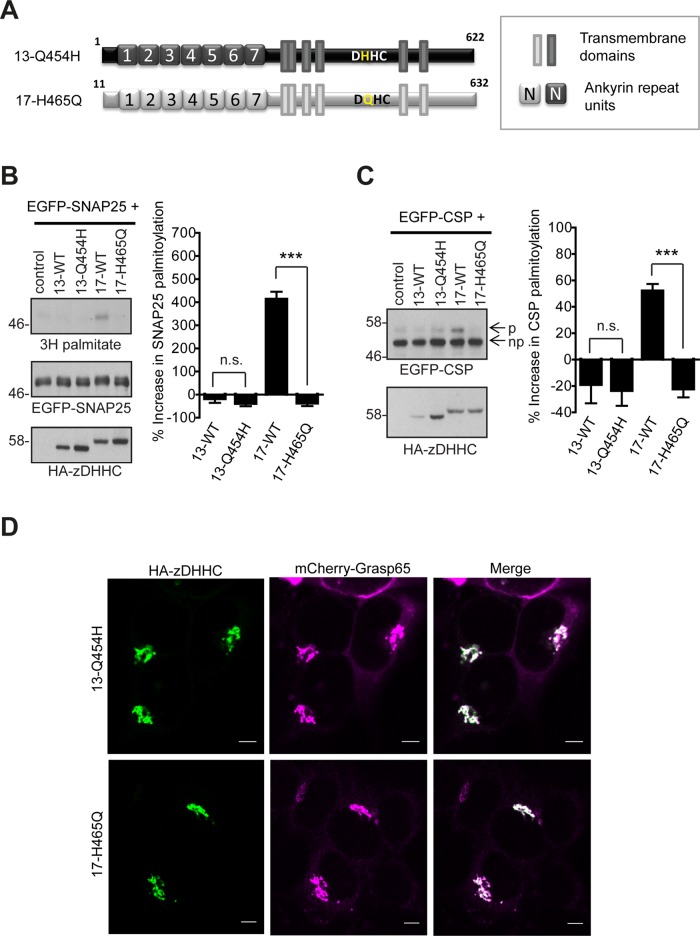
FIGURE 8:
Effect of the DQHC/DHHC motif in zDHHC13 and zDHHC17 on S-acylation of SNAP25 and CSP. (A) Schematic diagram of zDHHC17/13 mutants used for S-acylation assays. HEK293T cells were cotransfected with either EGFP-SNAP25 or EGFP-CSP and the indicated HA-tagged zDHHC construct. EGFP-SNAP25, EGFP-CSP, and HA-zDHHC enzymes were detected by immunoblotting with GFP and HA antibodies, respectively, and incorporation of radiolabel was detected with the aid of a Kodak Biomax Transcreen LE. Positions of molecular weight markers are shown on the left. SNAP25 palmitoylation was assessed by the amount of 3H-palmitic acid incorporated (after metabolic labeling) relative to protein level (B), and CSP palmitoylation was assessed after separation of its palmitoylated (p) and nonpalmitoylated (np) forms by SDS–PAGE of cell lysates by calculating the ratio of palmitoylated EGFP-CSP (p) to the total protein (p + np). (C) Percentage increase in palmitoylation (vs. control) was quantified (SNAP25, n = 4; CSP, n = 6; error bars, SEM). Statistical analysis was performed using a one-way ANOVA and Tukey posttest (n.s., p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). (D) Subcellular distribution of zDHHC17/13 mutants was assessed by cotransfection with Golgi marker GRASP65 (mCherry construct) and immunofluorescence using an HA antibody. Scale bars, 5 μm.