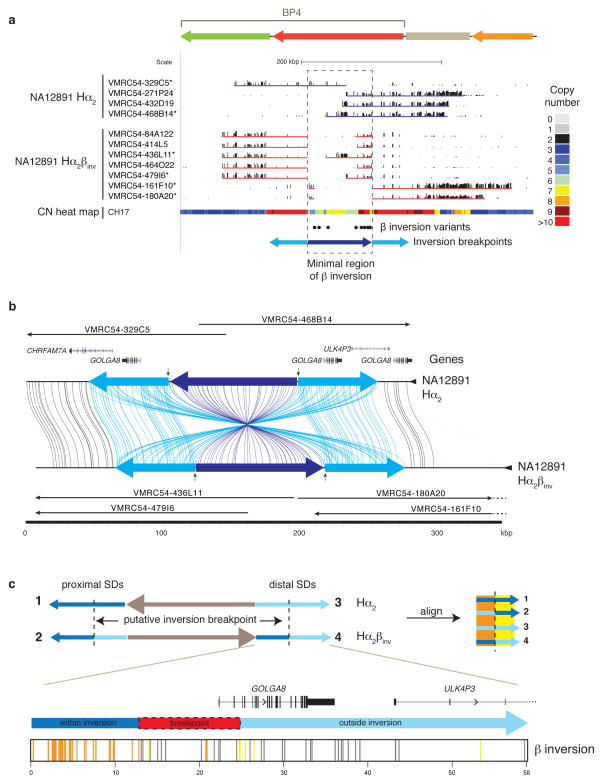
Figure 2. Sequence refinement of β inversion breakpoints.
(a) A 210 kbp β inversion was identified, validated, and sequenced using the VMRC54 BAC library (NA12891 individual). Illumina-generated sequences of clones spanning the BP4 CNPβ were mapped to human reference GRCh37. Clones sequenced using PacBio are indicated with asterisks. The copy number (CN) heat map shows the total diploid CN of a region in the CH17 hydatidiform mole cell line. The locations of the β inversion haplotype-tagging variants are pictured as dots. The blue arrows represent the BP4 CNPβ (dark blue) with the flanking 58 kbp inverted SDs (light blue). (b) Homologous sequences of clones, generated using PacBio and assembled into sequence contigs, are connected with colored lines between the direct (Hα2) and inverted (Hα2βinv) haplotypes from NA12891 using Miropeats55. Vertical arrows indicate the minimal inversion breakpoints. (c) Homologous sequences (58 kbp) from the BP4 CNPβ flanking inverted SDs were aligned from multiple individuals (NA12891 and CH17) and haplotypes (β direct: SDs 1 and 3, and β inverse: SDs 2 and 4; see Supplementary Figure 5 for a more detailed alignment) and variant sites compared. Variant positions showing signatures of being within or outside of the β inversion breakpoints are indicated as colored lines under the picture of the distal β inverse SD including: within the inversion (orange; consensus of SDs 1 & 4 and SDs 2 & 3), outside the inversion (yellow; consensus of SDs 1 & 2 and SDs 3 & 4), and gene conversion (gray; consensus of SDs 1 & 3 and SDs 2& 4). The inversion breakpoint, refined to a region in which we observe a transition from orange to yellow lines, is highlighted with a dash-outlined red box.