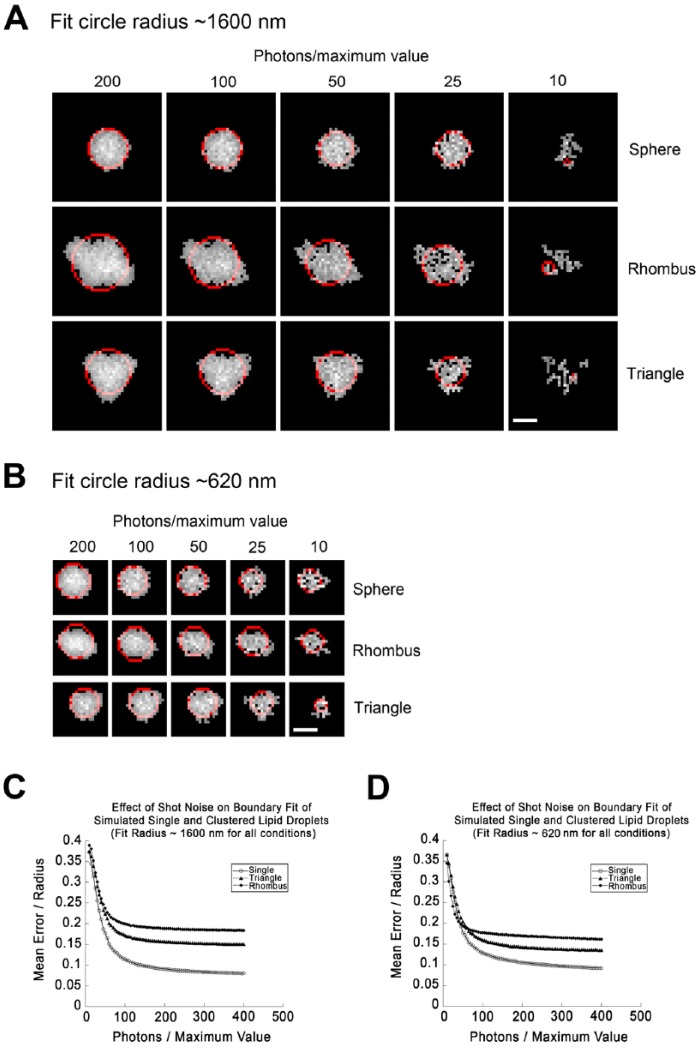
Figure 3.
Effects of increased noise on the ability to distinguish simulated spherical lipid droplets (LDs) from clusters. (A) LDs were simulated as described for Figure 2 to give a diameter for the fit circle of roughly 1600 nm. This corresponded to an initial simulated size of 2300 nm for a single LD, 1300 nm for each LD arranged in a triangle, and 1100 nm for each LD arranged in a rhombus. LDs were subjected to simulated photon shot noise with the brightest simulated pixel set to the indicated number of photons/pixel. Noised images were then thresholded to their full width half maximum (FWHM) and the boundary extracted and fit to a circle (red). Note that images degrade significantly as smaller numbers of simulated photons are sampled. (B) LDs simulated as in (A), but with sizes of LDs set to give a fit circle diameter near 620 nm for each geometry. This corresponds to an initial radius prior to processing of 400 nm for single LDs and 200 nm for each LD arranged in a rhombus or triangle. (C) Fit error (SDBFC; standard-deviation best-fit-circle) as a function of fit circle radius for single LDs, clusters of three arranged in a triangle, or clusters of four arranged in a rhombus with an overlap of 20% created and processed such that the fit circle diameter approximates 16 pixels (1600 nm) exactly as described for (A). Each point represents the mean value of 1000 simulations. Simulations were not included in the average if the structure was broken into more than one contiguous area. This was <5% of total simulations at 40 photons/maximum value for all geometries, but rejections for this reason rapidly increased at higher noise levels. (D) Effect of shot noise on fit error exactly as in (C) but with a fit circle diameter approximating 600 nm. Scale, 10 simulated pixels (1.0 simulated µm).