Abstract
Motivated by recent work on studying massive imaging data in various neuroimaging studies, we propose a novel spatially varying coefficient model (SVCM) to capture the varying association between imaging measures in a three-dimensional (3D) volume (or 2D surface) with a set of covariates. Two stylized features of neuorimaging data are the presence of multiple piecewise smooth regions with unknown edges and jumps and substantial spatial correlations. To specifically account for these two features, SVCM includes a measurement model with multiple varying coefficient functions, a jumping surface model for each varying coefficient function, and a functional principal component model. We develop a three-stage estimation procedure to simultaneously estimate the varying coefficient functions and the spatial correlations. The estimation procedure includes a fast multiscale adaptive estimation and testing procedure to independently estimate each varying coefficient function, while preserving its edges among different piecewise-smooth regions. We systematically investigate the asymptotic properties (e.g., consistency and asymptotic normality) of the multiscale adaptive parameter estimates. We also establish the uniform convergence rate of the estimated spatial covariance function and its associated eigenvalues and eigenfunctions. Our Monte Carlo simulation and real data analysis have confirmed the excellent performance of SVCM.
Keywords: Asymptotic normality, Functional principal component analysis, Jumping surface model, Kernel, Spatial varying coefficient model, Wald test
1 Introduction
The aims of this paper are to develop a spatially varying coefficient model (SVCM) to delineate association between massive imaging data and a set of covariates of interest, such as age, and to characterize the spatial variability of the imaging data. Examples of such imaging data include T1 weighted magnetic resonance imaging (MRI), functional MRI, and diffusion tensor imaging, among many others (Friston, 2007; Thompson and Toga, 2002; Mori, 2002; Lazar, 2008). In neuroimaging studies, following spatial normalization, imaging data usually consists of data points from different subjects (or scans) at a large number of locations (called voxels) in a common 3D volume (without loss of generality), which is called a template. We assume that all imaging data have been registered to a template throughout the paper.
To analyze such massive imaging data, researchers face at least two main challenges. The first one is to characterize varying association between imaging data and covariates, while preserving important features, such as edges and jumps, and the shape and spatial extent of effect images. Due to the physical and biological reasons, imaging data are usually expected to contain spatially contiguous regions or effect regions with relatively sharp edges (Chumbley et al., 2009; Chan and Shen, 2005; Tabelow et al., 2008a, b). For instance, normal brain tissue can generally be classified into three broad tissue types including white matter, gray matter, and cerebrospinal fluid. These three tissues can be roughly separated by using MRI due to their imaging intensity differences and relatively intensity homogeneity within each tissue. The second challenge is to characterize spatial correlations among a large number of voxels, usually in the tens thousands to millions, for imaging data. Such spatial correlation structure and variability are important for achieving better prediction accuracy, for increasing the sensitivity of signal detection, and for characterizing the random variability of imaging data across subjects (Cressie and Wikle, 2011; Spence et al., 2007).
There are two major statistical methods including voxel-wise methods and multiscale adaptive methods for addressing the first challenge. Conventional voxel-wise approaches involve in Gaussian smoothing imaging data, independently fitting a statistical model to imaging data at each voxel, and generating statistical maps of test statistics and p-values (Lazar, 2008; Worsley et al., 2004). As shown in Chumbley et al. (2009) and Li et al. (2011), voxel-wise methods are generally not optimal in power since it ignores the spatial information of imaging data. Moreover, the use of Gaussian smoothing can blur the image data near the edges of the spatially contiguous regions and thus introduce substantial bias in statistical results (Yue et al., 2010).
There is a great interest in the development of multiscale adaptive methods to adaptively smooth neuroimaging data, which is often characterized by a high noise level and a low signal-to-noise ratio (Tabelow et al., 2008a, b; Polzehl et al., 2010; Li et al., 2011; Qiu, 2005, 2007). Such multiscale adaptive methods not only increase signal-to-noise ratio, but also preserve important features (e.g., edge) of imaging data. For instance, in Polzehl and Spokoiny (2000, 2006), a novel propagation-separation approach was developed to adaptively and spatially smooth a single image without explicitly detecting edges. Recently, there are a few attempts to extend those adaptive smoothing methods to smoothing multiple images from a single subject (Tabelow et al., 2008a, b; Polzehl et al., 2010). In Li et al. (2011), a multiscale adaptive regression model, which integrates the propagation-separation approach and voxel-wise approach, was developed for a large class of parametric models.
There are two major statistical models, including Markov random fields and low rank models, for addressing the second challenge. The Markov random field models explicitly use the Markov property of an undirected graph to characterize spatial dependence among spatially connected voxels (Besag, 1986; Li, 2009). However, it can be restrictive to assume a specific type of spatial correlation structure, such as Markov random fields, for very large spatial data sets besides its computational complexity (Cressie and Wikle, 2011). In spatial statistics, low rank models, also called spatial random effects models, use a linear combination of ‘known’ spatial basis functions to approximate spatial dependence structure in a single spatial map (Cressie and Wikle, 2011). The low rank models have a close connection with the functional principal component analysis model for characterizing spatial correlation structure in multiple images, in which spatial basis functions are directly estimated (Zipunnikov et al., 2011; Ramsay and Silverman, 2005; Hall et al., 2006).
The goal of this article is to develop SVCM and its estimation procedure to simultaneously address the two challenges discussed above. SVCM has three features: piecewise smooth, spatially correlated, and spatially adaptive, while its estimation procedure is fast, accurate and individually updated. Major contributions of the paper are as follows.
Compared with the existing multiscale adaptive methods, SVCM first integrates a jumping surface model to delineate the piecewise smooth feature of raw and effect images and the functional principal component model to explicitly incorporate the spatial correlation structure of raw imaging data.
A comprehensive three-stage estimation procedure is developed to adaptively and spatially improve estimation accuracy and capture spatial correlations.
Compared with the existing methods, we use a fast and accurate estimation method to independently smooth each of effect images, while consistently estimating their standard deviation images.
We systematically establish consistency and asymptotic distribution of the adaptive parameter estimators under two different scenarios including piecewise-smooth and piecewise-constant varying coefficient functions. In particular, we introduce several adaptive boundary conditions to delineate the relationship between the amount of jumps and the sample size. Our conditions and theoretical results differ substantially from those for the propagation-separation type methods (Polzehl and Spokoiny, 2000, 2006; Li et al., 2011).
The rest of this paper is organized as follows. In Section 2, we describe SVCM and its three-stage estimation procedure and establish the theoretical properties. In Section 3, we present a set of simulation studies with the known ground truth to examine the finite sample performance of the three-stage estimation procedure for SVCM. In Section 4, we apply the proposed methods in a real imaging dataset on attention deficit hyper-activity disorder (ADHD). In Section 5, we conclude the paper with some discussions. Technical conditions are given in Section 6. Proofs and additional results are given in a supplementary document.
2 Spatial Varying Coefficient Model with Jumping Discontinuities
2.1 Model Setup
We consider imaging measurements in a template and clinical variables (e.g., age, gender, and height) from n subjects. Let
 represent a 3D volume and d and d0, respectively, denote a point and the center of a voxel in
represent a 3D volume and d and d0, respectively, denote a point and the center of a voxel in
 . Let
. Let
 be the union of all centers d0 in
be the union of all centers d0 in
 and ND equal the number of voxels in
and ND equal the number of voxels in
 . Without loss of generality,
. Without loss of generality,
 is assumed to be a compact set in R3. For the i-th subject, we observe an m × 1 vector of imaging measures yi(d0) at d0 ∈
is assumed to be a compact set in R3. For the i-th subject, we observe an m × 1 vector of imaging measures yi(d0) at d0 ∈
 , which leads to an mND × 1 vector of measurements across
, which leads to an mND × 1 vector of measurements across
 , denoted by
, denoted by
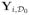 = {yi(d0) : d0 ∈
= {yi(d0) : d0 ∈
 }. For notational simplicity, we set m = 1 and consider a 3D volume throughout the paper.
}. For notational simplicity, we set m = 1 and consider a 3D volume throughout the paper.
The proposed spatial varying coefficient model (SVCM) consists of three components: a measurement model, a jumping surface model, and a functional component analysis model. The measurement model characterizes the association between imaging measures and covariates and is given by
| (1) |
where xi = (xi1, …, xip)T is a p × 1 vector of covariates, β(d) = (β1(d), …, βp(d))T is a p × 1 vector of coefficient functions of d, ηi(d) characterizes individual image variations from
, and εi(d) are measurement errors. Moreover, {ηi(d) : d ∈
 } is a stochastic process indexed by d ∈
} is a stochastic process indexed by d ∈
 that captures the within-image dependence. We assume that they are mutually independent and ηi(d) and εi(d) are independent and identical copies of SP(0, Ση) and SP(0, Σε), respectively, where SP(μ, Σ) denotes a stochastic process vector with mean function μ(d) and covariance function Σ(d, d′). Moreover, εi(d) and εi(d′) are independent for d ≠ d′ and thus Σε(d, d′) = 0 for d ≠ d′. Therefore, the covariance function of {yi(d) : d ∈
that captures the within-image dependence. We assume that they are mutually independent and ηi(d) and εi(d) are independent and identical copies of SP(0, Ση) and SP(0, Σε), respectively, where SP(μ, Σ) denotes a stochastic process vector with mean function μ(d) and covariance function Σ(d, d′). Moreover, εi(d) and εi(d′) are independent for d ≠ d′ and thus Σε(d, d′) = 0 for d ≠ d′. Therefore, the covariance function of {yi(d) : d ∈
 }, conditioned on xi, is given by
}, conditioned on xi, is given by
| (2) |
The second component of the SVCM is a jumping surface model for each of {βj(d) : d ∈
 }j≤p. Imaging data {yi(d0) : d0 ∈
}j≤p. Imaging data {yi(d0) : d0 ∈
 } can usually be regarded as a noisy version of a piecewise-smooth function of d ∈
} can usually be regarded as a noisy version of a piecewise-smooth function of d ∈
 with jumps or edges. In many neuroimaging data, those jumps or edges often reflect the functional and/or structural changes, such as white matter and gray matter, across the brain. Therefore, the varying function {βj(d) : d ∈
with jumps or edges. In many neuroimaging data, those jumps or edges often reflect the functional and/or structural changes, such as white matter and gray matter, across the brain. Therefore, the varying function {βj(d) : d ∈
 } in model (1) may inherit the piecewise-smooth feature from imaging data for j = 1, …, p, but allows to have different jumps and edges. Specially, we make the following assumptions.
} in model (1) may inherit the piecewise-smooth feature from imaging data for j = 1, …, p, but allows to have different jumps and edges. Specially, we make the following assumptions.
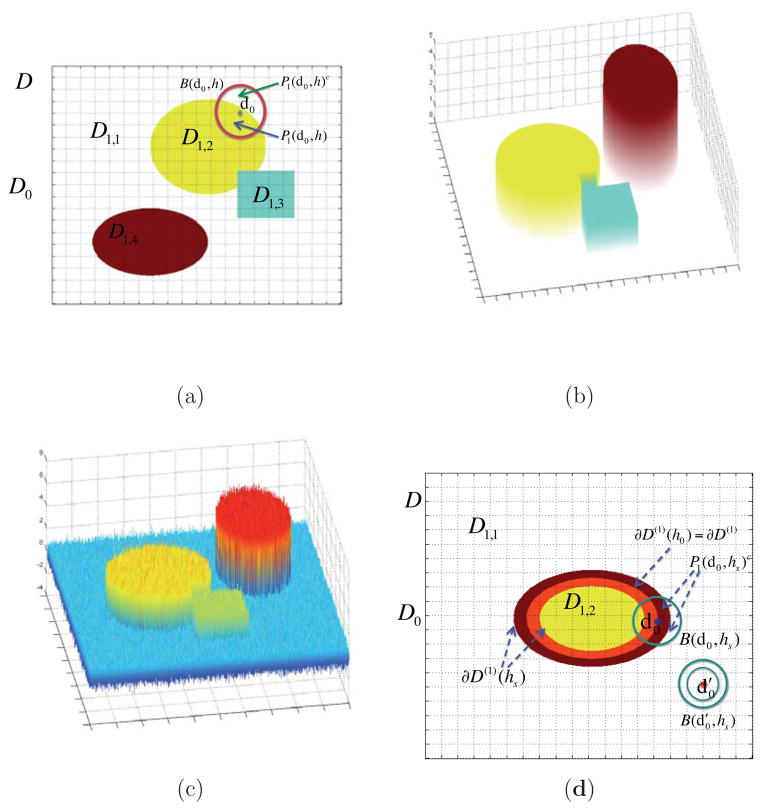
(i) (Disjoint Partition) There is a finite and disjoint partition {
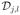 : l = 1, ···, Lj} of
: l = 1, ···, Lj} of
 such that each
such that each
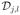 is a connected region of
is a connected region of
 and its interior, denoted by
, is nonempty, where Lj is a fixed, but unknown integer. See Figure 1 (a), (b), and (d) for an illustration.
and its interior, denoted by
, is nonempty, where Lj is a fixed, but unknown integer. See Figure 1 (a), (b), and (d) for an illustration.(ii) (Piecewise Smoothness) βj(d) is a smooth function of d within each for l = 1, …, Lj, but βj(d) is discontinuous on , which is the union of the boundaries of all
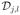 . See Figure 1 (b) for an illustration.
. See Figure 1 (b) for an illustration.(iii) (Local Patch) For any d0 ∈
 and h > 0, let B(d0, h) be an open ball of d0 with radius h and Pj(d0, h) a maximal path-connected set in B(d0, h), in which βj(d) is a smooth function of d. Assume that Pj(d0, h), which will be called a local patch, contains an open set. See Figure 1 for a graphical illustration.
and h > 0, let B(d0, h) be an open ball of d0 with radius h and Pj(d0, h) a maximal path-connected set in B(d0, h), in which βj(d) is a smooth function of d. Assume that Pj(d0, h), which will be called a local patch, contains an open set. See Figure 1 for a graphical illustration.
Figure 1.
Illustration of a jumping surface model for β1(d) and boundary sets over a two-dimensional region D: (a)
 ,
,
 , a disjoint partition of
, a disjoint partition of
 as the union of four disjoint regions with white, yellow, blue green, and red representing
as the union of four disjoint regions with white, yellow, blue green, and red representing
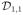 ,
,
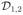 ,
,
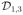 , and
, and
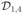 , a representative voxel d0 ∈
, a representative voxel d0 ∈
 , an open ball of d0, B(d0, h), a maximal path-connected set P1(d0, h), and P1(d0, h)c; (b) three-dimensional shaded surface of true {β1(d): d ∈
, an open ball of d0, B(d0, h), a maximal path-connected set P1(d0, h), and P1(d0, h)c; (b) three-dimensional shaded surface of true {β1(d): d ∈
 } map; (c) three-dimensional shaded surface of estimated {β̂1(d0): d0 ∈
} map; (c) three-dimensional shaded surface of estimated {β̂1(d0): d0 ∈
 } map; and (d)
} map; and (d)
 ,
,
 , a disjoint partition of
, a disjoint partition of
 =
=
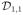 ∪
∪
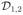 , ∂D(1)(h0) ⊂ ∂D(1)(hs), two representative voxels d0 and
in
, ∂D(1)(h0) ⊂ ∂D(1)(hs), two representative voxels d0 and
in
 , two open balls of
, an open ball of d0 ∈ ∂D(1)(hs) ∩
, two open balls of
, an open ball of d0 ∈ ∂D(1)(hs) ∩
 , B(d0, hs), and P1(d0, hs)c.
, B(d0, hs), and P1(d0, hs)c.
The jumping surface model can be regarded as a generalization of various models for delineating changes at unknown location (or time). See, for example, Khodadadi and Asgharian (2008) for an annotated bibliography of change point problem and regression. The disjoint partition and piecewise smoothness assumptions characterize the shape and smoothness of βj(d) in
 , whereas the local patch assumption primarily characterizes the local shape of βj(d) at each voxel d0 ∈
, whereas the local patch assumption primarily characterizes the local shape of βj(d) at each voxel d0 ∈
 across different scales (or radii). For
, there exists a radius h(d0) such that
. In this case, for h ≤ h(d0), we have Pj(d0, h) = B(d0, h) and Pj(d0, h)c = ∅, whereas Pj(d0, h)c may not equal the empty set for large h since B(d0, h) may cross different
. For d0 ∈ ∂
across different scales (or radii). For
, there exists a radius h(d0) such that
. In this case, for h ≤ h(d0), we have Pj(d0, h) = B(d0, h) and Pj(d0, h)c = ∅, whereas Pj(d0, h)c may not equal the empty set for large h since B(d0, h) may cross different
. For d0 ∈ ∂
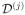 ∩
∩
 , Pj(d0, h)c ≠ ∅ for all h > 0. Since Pj(d0, h) contains an open set for any h > 0, it eliminates the case of d0 being an isolated point. See Figure 1 (a) and (d) for an illustration.
, Pj(d0, h)c ≠ ∅ for all h > 0. Since Pj(d0, h) contains an open set for any h > 0, it eliminates the case of d0 being an isolated point. See Figure 1 (a) and (d) for an illustration.
The last component of the SVCM is a functional principal component analysis model for ηi(d). Let λ1 ≥ λ2 ≥ …≥ 0 be ordered values of the eigenvalues of the linear operator determined by Ση with and the ψl(d)s’ be the corresponding orthonormal eigenfunctions (or principal components) (Li and Hsing, 2010; Hall et al., 2006). Then, Ση admits the spectral decomposition:
| (3) |
The eigenfunctions ψl(d) form an orthonormal basis on the space of square-integrable functions on
 , and ηi(d) admits the Karhunen-Loeve expansion as follows:
, and ηi(d) admits the Karhunen-Loeve expansion as follows:
| (4) |
where ξi,l =
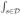 ηi(s)ψl(s)d
ηi(s)ψl(s)d
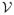 (s) is referred to as the l-th functional principal component score of the ith subject, in which d
(s) is referred to as the l-th functional principal component score of the ith subject, in which d
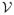 (s) denotes the Lebesgue measure. The ξi,l are uncorrelated random variables with E(ξi,l) = 0 and E(ξi,lξi,k) = λl1(l = k). If λl ≈ 0 for l ≥ LS + 1, then model (1) can be approximated by
(s) denotes the Lebesgue measure. The ξi,l are uncorrelated random variables with E(ξi,l) = 0 and E(ξi,lξi,k) = λl1(l = k). If λl ≈ 0 for l ≥ LS + 1, then model (1) can be approximated by
| (5) |
In (5), since ξi,l are random variables and ψl(d) are ‘unknown’ but fixed basis functions, it can be regarded as a varying coefficient spatial mixed effects model. Therefore, model (5) is a mixed effects representation of model (1).
Model (5) differs significantly from other models in the existing literature. Most varying coefficient models assume some degrees of smoothness on varying coefficient functions, while they do not model the within-curve dependence (Wu et al., 1998). See Fan and Zhang (2008) for a comprehensive review of varying coefficient models. Most spatial mixed effects models in spatial statistics assume that spatial basis functions are known and regression coefficients do not vary across d (Cressie and Wikle, 2011). Most functional principal component analysis models focus on characterizing spatial correlation among multiple observed functions when
 ∈ R1 (Zipunnikov et al., 2011; Ramsay and Silverman, 2005; Hall et al., 2006).
∈ R1 (Zipunnikov et al., 2011; Ramsay and Silverman, 2005; Hall et al., 2006).
2.2 Three-stage Estimation Procedure
We develop a three-stage estimation procedure as follows. See Figure 2 for a schematic overview of SVCM.
Figure 2.
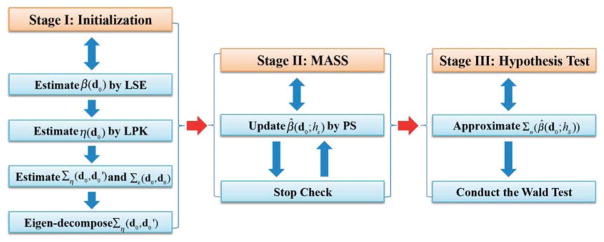
A schematic overview of the three stages of SVCM: Stage (I) is the initialization step, Stage (II) is the Multiscale Adaptive and Sequential Smoothing (MASS) method, and Stage (III) is the hypothesis test.
Stage (I): Calculate the least squares estimate of β(d0), denoted by β̂(d0), across all voxels in
 , and estimate {Σε(d0, d0) : d0 ∈
, and estimate {Σε(d0, d0) : d0 ∈
 }, {Ση(d, d′) : (d, d′) ∈
}, {Ση(d, d′) : (d, d′) ∈
 } and its eigenvalues and eigenfunctions.
} and its eigenvalues and eigenfunctions.Stage (II): Use the propagation-seperation method to adaptively and spatially smooth each component of β̂(d0) across all d0 ∈
 .
.Stage (III): Approximate the asymptotic covariance matrix of the final estimate of β(d0) and calculate test statistics across all voxels d0 ∈
 .
.
This is more refined idea than the two-stage procedure proposed in Fan and Zhang (1999, 2002).
2.2.1 Stage (I)
Stage (I) consists of four steps.
Step (I.1) is to calculate the least squares estimate of β(d0), which equals
across all voxels d0 ∈
 , where
, in which a⊗2 = aaT for any vector a. See Figure 1 (c) for a graphical illustration of {β̂(d0) : d0 ∈
, where
, in which a⊗2 = aaT for any vector a. See Figure 1 (c) for a graphical illustration of {β̂(d0) : d0 ∈
 }.
}.
Step (I.2) is to estimate ηi(d) for all d ∈
 . We employ the local linear regression technique to estimate all individual functions ηi(d). Let ∂dηi(d) = ∂ηi(d)/∂d, Ci(d) = (ηi(d), h∂dηi(d)T)T, and zh(dm − d) = (1, (dm,1 − d1)/h, (dm,2 − d2)/h, (dm,3 − d3)/h)T, where d = (d1, d2, d3)T and dm = (dm,1, dm,2, dm,3)T ∈
. We employ the local linear regression technique to estimate all individual functions ηi(d). Let ∂dηi(d) = ∂ηi(d)/∂d, Ci(d) = (ηi(d), h∂dηi(d)T)T, and zh(dm − d) = (1, (dm,1 − d1)/h, (dm,2 − d2)/h, (dm,3 − d3)/h)T, where d = (d1, d2, d3)T and dm = (dm,1, dm,2, dm,3)T ∈
 . We use Taylor series expansion to expand ηi(dm) at d leading to
. We use Taylor series expansion to expand ηi(dm) at d leading to
We develop an algorithm to estimate Ci(d) as follows. Let Kloc(·) be a univariate kernel function and be the rescaled kernel function with a bandwidth h. For each i, we estimate Ci(d) by minimizing the weighted least squares function given by
where . It can be shown that
| (6) |
Let R̂i = (ri(d0) : d0 ∈
 ) be an ND × 1 vector of estimated residuals and notice that η̂i(d) is the first component of Ci(d). Then, we have
) be an ND × 1 vector of estimated residuals and notice that η̂i(d) is the first component of Ci(d). Then, we have
| (7) |
where Si is an ND × ND smoothing matrix (Fan and Gijbels, 1996). We pool the data from all n subjects and select the optimal bandwidth h, denoted by h̃, by minimizing the generalized cross-validation (GCV) score given by
| (8) |
where ID is an ND × ND identity matrix. Based on h̃, we can use (7) to estimate ηi(d) for all i.
Step (I.3) is to estimate Ση(d, d′) and Σε(d0, d0). Let
be estimated residuals for i = 1, …, n and d0 ∈
 . We estimate Σε(d0, d0) by
. We estimate Σε(d0, d0) by
| (9) |
and Ση(d, d′) by the sample covariance matrix:
| (10) |
Step (I.4) is to estimate the eigenvalue-eigenfunction pairs of Ση by using the singular value decomposition. Let V = [η̂1, ···, η̂n] be an ND × n matrix. Since n is much smaller than ND, we can easily calculate the eigenvalue-eigenvector pairs of the n × n matrix VTV, denoted by {(λ̂ i, ξ̂i) : i = 1, ···, n}. It can be shown that {(λ̂i, Vξ̂i) : i = 1, ···, n} are the eigenvalue-eigenvector pairs of the ND × ND matrix VVT. In applications, one usually considers large λ̂l values, while dropping small λ̂ls. It is common to choose a value of LS so that the cumulative eigenvalue is above a prefixed threshold, say 80% (Zipunnikov et al., 2011; Li and Hsing, 2010; Hall et al., 2006). Furthermore, the lth SPCA scores can be computed using
| (11) |
for l = 1, …, LS, where
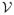 (dm) is the volume of voxel dm.
(dm) is the volume of voxel dm.
2.2.2 Stage (II)
Stage (II) is a multiscale adaptive and sequential smoothing (MASS) method. The key idea of MASS is to use the propagation-separation method (Polzehl and Spokoiny, 2000, 2006) to individually smooth each least squares estimate image {β̂j(d0) : d0 ∈
 } for j = 1, …, p. MASS starts with building a sequence of nested spheres with increasing bandwidths 0 = h0 < h1 < ··· < hS = r0 ranging from the smallest bandwidth h1 to the largest bandwidth hS = r0 for each d0 ∈
} for j = 1, …, p. MASS starts with building a sequence of nested spheres with increasing bandwidths 0 = h0 < h1 < ··· < hS = r0 ranging from the smallest bandwidth h1 to the largest bandwidth hS = r0 for each d0 ∈
 . At bandwidth h1, based on the information contained in {β̂(d0) : d0 ∈
. At bandwidth h1, based on the information contained in {β̂(d0) : d0 ∈
 }, we sequentially calculate adaptive weights
between voxels d0 and
, which depends on the distance ||d0 −d0|| and spacial similarity |β̂j(d0) − β̂j(d0)|, and update β̂j(d0; h1) for all d0 ∈
}, we sequentially calculate adaptive weights
between voxels d0 and
, which depends on the distance ||d0 −d0|| and spacial similarity |β̂j(d0) − β̂j(d0)|, and update β̂j(d0; h1) for all d0 ∈
 for j = 1, ···, p. At bandwidth h2, we repeat the same process using {β̂(d0; h1) : d0 ∈
for j = 1, ···, p. At bandwidth h2, we repeat the same process using {β̂(d0; h1) : d0 ∈
 } to compute spatial similarities. In this way, we can sequentially determine
and β̂j(d0; hs) for each component of β(d0) as the bandwidth ranges from h1 to hS = r0. Moreover, as shown below, we have found a simple way of calculating the standard deviation of β̂j(d0; hs).
} to compute spatial similarities. In this way, we can sequentially determine
and β̂j(d0; hs) for each component of β(d0) as the bandwidth ranges from h1 to hS = r0. Moreover, as shown below, we have found a simple way of calculating the standard deviation of β̂j(d0; hs).
MASS consists of three steps including (II.1) an initialization step, (II.2) a sequentially adaptive estimation step, and (II.3) a stop checking step, each of which involves in the specification of several parameters. Since propagation-separation and the choice of their associated parameters have been discussed in details in Polzehl et al. (2010) and Li et al. (2011), we briefly mention them here for the completeness. In the initialization step (II.1), we take a geometric series { : s = 1, …, S} of radii with h0 = 0, where ch > 1, say ch = 1.10. We suggest relatively small ch to prevent incorporating too many neighboring voxels.
In the sequentially adaptive estimation step (II.2), starting from s = 1 and h1 = ch, at step s, we compute spatial adaptive locally weighted average estimate β̂j(d0; hs) based on {β̂j(d0) : d0 ∈
 } and {β̂j(d0; hs−1) : d ∈
} and {β̂j(d0; hs−1) : d ∈
 }, where β̂j(d0; h0) = β̂j(d0). Specifically, for each j, we construct a weighted quadratic function
}, where β̂j(d0; h0) = β̂j(d0). Specifically, for each j, we construct a weighted quadratic function
| (12) |
where ωj(d0, dm; hs), which will be defined below, characterizes the similarity between β̂j(dm; hs−1) and β̂j(d0; hs−1). We then calculate
| (13) |
where ω̃j(d0, dm; hs) = ωj(d0, dm; hs)/
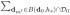 ωj(d0, dm′ ; hs).
ωj(d0, dm′ ; hs).
Let Σn(β̂j(d0; hs)) be the asymptotic variance of β̂j(d0; hs). For βj(d0), we compute the similarity between voxels d0 and , denoted by , and the adaptive weight , which are, respectively, defined as
| (14) |
where Kst(u) is a nonnegative kernel function with compact support, Cn is a tuning parameter depending on n, and || · ||2 denotes the Euclidean norm of a vector.
The weights give less weight to the voxel that is far from the voxel d0. The weights Kst(u) downweight the voxels with large , which indicates a large difference between and β̂j(d0; hs−1). In practice, we set Kloc(u) = (1 − u)+. Although different choices of Kst(·) have been suggested in the propagation-separation method (Polzehl and Spokoiny, 2000, 2006; Polzehl et al., 2010; Li et al., 2011), we have tested these kernel functions and found that Kst(u) = exp(−u) performs reasonably well. Another good choice of Kst(u) is min(1, 2(1 − u))+. Moreover, theoretically, as shown in Scott (1992) and Fan (1993), they have examined the efficiency of different kernels for weighted least squares estimators, but extending their results to the propagation-separation method needs some further investigation.
The scale Cn is used to penalize the similarity between any two voxels d0 and in a similar manner to bandwidth, and an appropriate choice of Cn is crucial for the behavior of the propagation-separation method. As discussed in (Polzehl and Spokoiny, 2000, 2006), a propagation condition independent of the observations at hand can be used to specify Cn. The basic idea of the propagation condition is that the impact of the statistical penalty in should be negligible under a homogeneous model βj(d) ≡ constant yielding almost free smoothing within homogeneous regions. However, we take an alternative approach to choose Cn here. Specifically, a good choice of Cn should balance between the sensitivity and specificity of MASS. Theoretically, as shown in Section 2.3, Cn should satisfy Cn/n = o(1) and . We choose based on our experiments, where is the upper a-percentile of the -distribution.
We now calculate Σn(β̂j(d0; hs)). By treating the weights ω̃j(d0, dm; hs) as ‘fixed’ constants, we can approximate Σn(β̂j(d0; hs)) by
| (15) |
where Cov(β̂j(dm), β̂j(dm′)) can be estimated by
| (16) |
in which ej,p is a p × 1 vector with the j-th element 1 and others 0. We will examine the consistency of approximation (15) later.
In the stop checking step (II.3), after the first iteration, we start to calculate a stopping criterion based on a normalized distance between β̂j(d0) and β̂j(d0; hs) given by
| (17) |
Then, we check whether β̂j(d0; hs) is in a confidence ellipsoid of β̂j(d0) given by {βj(d0) : D(β̂j(d0), βj(d0)) ≤ Cs}, where Cs is taken as in our implementation. If D(β̂j(d0), β̂j(d0; hs)) is greater than Cs, then we set β̂j(d0, hS) = β̂j(d0, hs−1) and s = S for the j-th component and voxel d0. If s = S for all components in all voxels, we stop. If D(β̂j(d0), β̂j(d0; hs)) ≤ Cs, then we set hs+1 = chhs, increase s by 1 and continue with the step (II.1). It should be noted that different components of β̂(d0; h) may stop at different bandwidths.
We usually set the maximal step S to be relatively small, say between 10 and 20, and thus each B(d0, hS) only contains a relatively small number of voxels. As S increases, the number of neighboring voxels in B(d0, hS) increases exponentially. It increases the chance of oversmoothing βj(d0) when d0 is near the edge of distinct regions. Moreover, in order to prevent oversmoothing βj(d0), we compare β̂j(d0; hs) with the least squares estimate β̂j(d0) and gradually decrease Cs with the number of iteration.
2.2.3 Stage (III)
Based on β̂(d0; hS), we can further construct test statistics to examine scientific questions associated with β(d0). For instance, such questions may compare brain structure across different groups (normal controls versus patients) or detect change in brain structure across time. These questions can be formulated as the linear hypotheses about β(d0) given by
| (18) |
where R1 is an r × k matrix of full row rank and b0 is an r × 1 specified vector. We use the Wald test statistic
| (19) |
for problem (18), where Σn(β̂(d0; hS)) is the covariance matrix of β̂(d0; hS).
We propose an approximation of Σn(β̂(d0; hS)). According to (13), we know that
where a ∘ b denotes the Hadamard product of matrices a and b and ω̃(d0, dm; h) is a p × 1 vector determined by the weights ω̃j(d0, dm; h) in Stage II. Let Jp be the p2 × p selection matrix (Liu, 1999). Therefore, Σn(β̂(d0; hS)) can be approximated by
2.3 Theoretical Results
We systematically investigate the asymptotic properties of all estimators obtained from the three-stage estimation procedure. Throughout the paper, we only consider a finite number of iterations and bounded r0 for MASS, since a brain volume is always bounded. Without otherwise stated, we assume that op(1) and Op(1) hold uniformly across all d in either
 or
or
 throughout the paper. Moreover, the sample size n and the number of voxels ND are allowed to diverge to infinity. We state the following theorems, whose detailed assumptions and proofs can be found in Section 6 and a supplementary document.
throughout the paper. Moreover, the sample size n and the number of voxels ND are allowed to diverge to infinity. We state the following theorems, whose detailed assumptions and proofs can be found in Section 6 and a supplementary document.
Let β*(d0) = (β1*(d0), …, βp*(d0))T be the true value of β(d0) at voxel d0. We first establish the uniform convergence rate of {β̂(d0) : d0 ∈
 }.
}.
Theorem 1
Under assumptions (C1)–(C4) in Section 6, as n → ∞, we have
(i) for any d0 ∈
 , where →L denotes convergence in distribution;
, where →L denotes convergence in distribution;(ii)
Remark 1
Theorem 1 (i) just restates a standard asymptotic normality of the least squares estimate of β(d0) at any given voxel d0 ∈
 . Theorem 1 (ii) states that the maximum of ||β̂(d0) − β*(d0)||2 across all d0 ∈
. Theorem 1 (ii) states that the maximum of ||β̂(d0) − β*(d0)||2 across all d0 ∈
 is at the order of
. If log(1 + ND) is relatively small compared with n, then the estimation errors converge uniformly to zero in probability. In practice, ND is determined by imaging resolution and its value can be much larger than the sample size. For instance, in most applications, ND can be as large as 1003 and log(1 + ND) is around 15. In a study with several hundreds subjects, n−1 log(1 + ND) can be relatively small.
is at the order of
. If log(1 + ND) is relatively small compared with n, then the estimation errors converge uniformly to zero in probability. In practice, ND is determined by imaging resolution and its value can be much larger than the sample size. For instance, in most applications, ND can be as large as 1003 and log(1 + ND) is around 15. In a study with several hundreds subjects, n−1 log(1 + ND) can be relatively small.
We next study the uniform convergence rate of Σ̂η and its associated eigenvalues and eigenfunctions. We also establish the uniform convergence of Σ̂ε(d0, d0).
Theorem 2
Under assumptions (C1)–(C8) in Section 6, we have the following results:
;
∫
 [ψ̂l(d) − ψ̂l(d)]2
d
[ψ̂l(d) − ψ̂l(d)]2
d
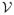 (d) = oP(1) and |λ̂l − λl| = op(1) for l = 1, …, E;
(d) = oP(1) and |λ̂l − λl| = op(1) for l = 1, …, E;;
where E will be described in assumption (C8) and ψ̂l(d) is the estimated eigenvector, computed from ψ̂l = Vξl.
Remark 2
Theorem 2 (i) and (ii) characterize the uniform weak convergence of Σ̂η(·, ·) and the convergence of ψ̂l (·) and λ̂l. These results can be regarded as an extension of Theorems 3.3–3.6 in Li and Hsing (2010), which established the uniform strong convergence rates of these estimates under a simple model. Specifically, in Li and Hsing (2010), they considered yi(d) = μ(d) + ηi(d) + εi(d) and assumed that μ(d) is twice differentiable. Another key difference is that in Li and Hsing (2010), they employed all cross products yi(d)yi(d′) for d ≠ d′ and then used the local polynomial kernel to estimate Ση(d, d′). In contrast, our approach is computationally simple and Σ̂η(d, d′) is positive definite. Theorem 2 (iii) characterizes the uniform weak convergence of Σ̂ε(d0, d0) across all voxels d0 ∈
 .
.
To investigate the asymptotic properties of β̂j(d0; hs), we need to characterize points close to and far from the boundary set ∂
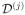 . For a given bandwidth hs, we first define hs-boundary sets:
. For a given bandwidth hs, we first define hs-boundary sets:
| (20) |
Thus, ∂
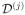 (hs) can be regarded as a band with radius hs covering the boundary set ∂
(hs) can be regarded as a band with radius hs covering the boundary set ∂
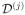 , while
contains all grid points within such band. It is easy to show that for a sequence of bandwidths h0 = 0 < h1 < ··· < hS, we have
, while
contains all grid points within such band. It is easy to show that for a sequence of bandwidths h0 = 0 < h1 < ··· < hS, we have
| (21) |
Therefore, for a fixed bandwidth hs, any point d0 ∈
 belongs to either
belongs to either
 \ ∂
\ ∂
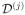 (hs) or ∂
(hs) or ∂
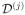 (hs). For each d0 ∈
(hs). For each d0 ∈
 \ ∂
\ ∂
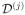 (hs), there exists one and only one
(hs), there exists one and only one
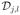 such that
such that
| (22) |
See Figure 1 (d) for an illustration.
We first investigate the asymptotic behavior of β̂j(d0; hs) when βj*(d) is piecewise constant. That is, βj*(d) is a constant in
and for any d′ ∈ ∂
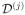 , there exists a
such that βj*(d) = βj*(d′). Let β̃j*(d0; hs) =
, there exists a
such that βj*(d) = βj*(d′). Let β̃j*(d0; hs) =
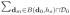 ω̃j(d0, dm; hs)βj*(dm) be the pseudo-true value of βj(d0) at scale hs in voxel d0. For all d0 ∈
ω̃j(d0, dm; hs)βj*(dm) be the pseudo-true value of βj(d0) at scale hs in voxel d0. For all d0 ∈
 \ ∂
\ ∂
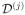 (hS), we have β̃j*(d0; hs) = βj*(d0) for all s ≤ S due to (22). In contrast, for d0 ∈ ∂
(hS), we have β̃j*(d0; hs) = βj*(d0) for all s ≤ S due to (22). In contrast, for d0 ∈ ∂
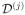 (hS), β̃j*(d0; hs) may vary from h0 to hS. In this case, we are able to establish several important theoretical results to characterize the asymptotic behavior of β̂(d0; hs) even when hS does not converge to zero. We need additional notation as follows:
(hS), β̃j*(d0; hs) may vary from h0 to hS. In this case, we are able to establish several important theoretical results to characterize the asymptotic behavior of β̂(d0; hs) even when hS does not converge to zero. We need additional notation as follows:
| (23) |
Theorem 3
Under assumptions (C1)–(C10) in Section 6 for piecewise constant {βj*(d) : d ∈
 }, we have the following results for all 0 ≤ s ≤ S:
}, we have the following results for all 0 ≤ s ≤ S:
;
;
;
converges in distribution to a normal distribution with mean zero and variance as n → ∞.
Remark 3
Theorem 3 shows that MASS has several important features for a piecewise constant function βj*(d). For instance, Theorem 3 (i) quantifies the maximum absolute difference (or bias) between the true value βj*(d0) and the pseudo true value β̃j*(d0; hs) across all d0 ∈
 for any s. Since β̃j*(d0; hs) − βj*(d0) = 0 for d0 ∈
for any s. Since β̃j*(d0; hs) − βj*(d0) = 0 for d0 ∈
 \ ∂
\ ∂
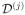 (hs), this result delineates the potential bias for voxels d0 in ∂
(hs), this result delineates the potential bias for voxels d0 in ∂
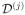 (hs).
(hs).
Theorem 3 (iv) ensures that is asymptotically normally distributed. Moreover, as shown in the supplementary document, is smaller than the asymptotic variance of the raw estimate β̂j(d0). As a result, MASS increases statistical power of testing H0(d0).
We now consider a much complex scenario when βj*(d) is piecewise smooth. In this case, β̃j*(d0; hs) may vary from h0 to hS for all voxels d0 ∈
 regardless whether d0 belongs to ∂
regardless whether d0 belongs to ∂
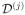 (hs) or not. We can establish important theoretical results to characterize the asymptotic behavior of β̂(d0; hs) only when
holds. We need some additional notation as follows:
(hs) or not. We can establish important theoretical results to characterize the asymptotic behavior of β̂(d0; hs) only when
holds. We need some additional notation as follows:
| (24) |
Theorem 4
Suppose assumptions (C1)–(C9) and (C11) in Section 6 hold for piecewise continuous {βj*(d) : d ∈
 }. For all 0 ≤ s ≤ S, we have the following results:
}. For all 0 ≤ s ≤ S, we have the following results:
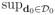 |β̃j*(d0; hs) − βj*(d0)| = Op(hs);
|β̃j*(d0; hs) − βj*(d0)| = Op(hs);;
.
converges in distribution to a normal distribution with mean zero and variance as n → ∞.
Remark 4
Theorem 4 characterizes several key features of MASS for a piecewise continuous function βj*(d). These results differ significantly from those for the piecewise constant case, but under weaker assumptions. For instance, Theorem 4 (i) quantifies the bias of the pseudo true value β̃j*(d0; hs) relative to the true value βj*(d0) across all d0 ∈
 for a fixed s. Even for voxels inside the smooth areas of βj*(d), the bias Op(hs) is still much higher than the standard bias at the rate of
due to the presence of
(Fan and Gijbels, 1996; Wand and Jones, 1995). If we set Kst(u) = 1(u ∈ [0, 1]) and βj*(d) is twice differentiable, then the bias of β̃j*(d0; hs) relative to βj*(d0) may be reduced to
. Theorem 4 (iv) ensures that
is asymptotically normally distributed. Moreover, as shown in the supplementary document,
is smaller than the asymptotic variance of the raw estimate β̂j(d0), and thus MASS can increase statistical power in testing H0(d0) even for the piecewise continuous case.
for a fixed s. Even for voxels inside the smooth areas of βj*(d), the bias Op(hs) is still much higher than the standard bias at the rate of
due to the presence of
(Fan and Gijbels, 1996; Wand and Jones, 1995). If we set Kst(u) = 1(u ∈ [0, 1]) and βj*(d) is twice differentiable, then the bias of β̃j*(d0; hs) relative to βj*(d0) may be reduced to
. Theorem 4 (iv) ensures that
is asymptotically normally distributed. Moreover, as shown in the supplementary document,
is smaller than the asymptotic variance of the raw estimate β̂j(d0), and thus MASS can increase statistical power in testing H0(d0) even for the piecewise continuous case.
3 Simulation Studies
In this section, we conducted a set of Monte Carlo simulations to compare MASS with voxel-wise methods from three different aspects. Firstly, we examine the finite sample performance of β̂(d0; hs) at different signal-to-noise ratios. Secondly, we examine the accuracy of the estimated eigenfunctions of Ση(d, d′). Thirdly, we assess both Type I and II error rates of the Wald test statistic. For the sake of space, we only present some selected results below and put additional simulation results in the supplementary document.
We simulated data at all 32,768 voxels on the 64 × 64 × 8 phantom image for n = 60 (or 80) subjects. At each d0 = (d0,1, d0,2, d0,3)T in
 , Yi(d0) was simulated according to
, Yi(d0) was simulated according to
| (25) |
where xi = (xi1, xi2, xi3)T, β(d0) = (β1(d0), β2(d0), β3(d0))T, and ε(d0) ~ N(0, 1) or χ(3)2 − 3, in which χ2(3) − 3 is a very skewed distribution. Furthermore, we set , where ξil are independently generated according to ξi1 ~ N(0, 0.6), ξi2 ~ N(0, 0.3), and ξi3 ~ N(0, 0.1), ψ1(d0) = 0.5 sin(2πd0,1/64), ψ2(d0) = 0.5 cos(2πd0,2/64), and . The first eigenfunction ψ1(d0) changes only along d0,1 direction, while it keeps constant in the other two directions. The other two eigenfunctions, ψ2(d0) and ψ3(d0), were chosen in a similar way (Figure 3). We set xi1 = 1 and generated xi2 independently from a Bernoulli distribution with success rate 0.5 and xi3 independently from the uniform distribution on [1, 2]. The covariates xi2 and xi3 were chosen to represent group identity and scaled age, respectively.
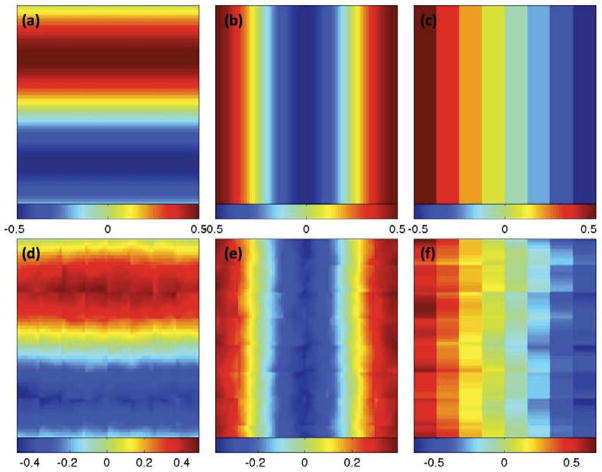
Figure 3.
Simulation results: a selected slice of (a) true ψ1(d); (b) true ψ2(d); (c) true ψ3(d); (d) ψ̂1(d); (e) ψ̂2(d); and (f) ψ̂3(d).
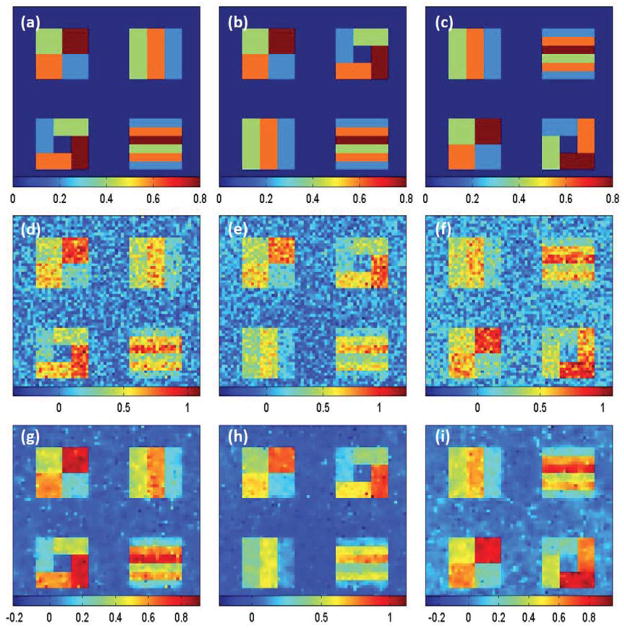
We chose different pattens for different βj(d) images in order to examine the finite sample performance of our estimation method under different scenarios. We set all the 8 slices along the coronal axis to be identical for each of βj(d) images. As shown in Figure 4, each slice of the three different βj(d) images has four different blocks and 5 different regions of interest (ROIs) with varying patterns and shape. The true values of βj(d) were varied from 0 to 0.8, respectively, and were displayed for all ROIs with navy blue, blue, green, orange and brown colors representing 0, 0.2, 0.4, 0.6, and 0.8, respectively.
Figure 4.
Simulation results: a selected slice of (a) true β1(d); (b) true β2(d); (c) true β3(d); (d) β̂1(d0); (e) β̂2(d0); (f) β̂3(d0); (g) β̂1(d0; h10); (h) β̂2(d0; h10); and (i) β̂3(d0; h10).
We fitted the SVCM model (1) with the same set of covariates to a simulated data set, and then applied the three-stage estimation procedure described in Section 2.2 to calculate adaptive parameter estimates across all pixels at 11 different scales. In MASS, we set hs = 1.1s for s = 0, …, S = 10. Figure 4 shows some selected slices of β̂(d0; hs) at s = 0 (middle panels) and s = 10 (lower panels). Inspecting Figure 4 reveals that all β̂j(d0; h10) outperform their corresponding β̂j(d0) in terms of variance and detected ROI patterns. Following the method described in Section 2.2, we estimated ηi(d) based on the residuals by using the local linear smoothing method and then calculate η̂i(d). Figure 3 shows some selected slices of the first three estimated eigenfunctions. Inspecting Figure 3 reveals that η̂i(d) are relatively close to the true eigenfunctions and can capture the main feature in the true eigenfunctions, which vary in one direction and are constant in the other two directions. However, we do observe some minor block effects, which may be caused by using the block smoothing method to estimate ηi(d).
Furthermore, for β̂(d0; hs), we calculated the bias, the empirical standard error (RMS), the mean of the estimated standard errors (SD), and the ratio of RMS over SD (RE) at each voxel of the five ROIs based on the results obtained from the 200 simulated data sets. For the sake of space, we only presented some selected results based on β̂3(d0) and β̂3(d0; h10) obtained from N(0, 1) distributed data with n = 60 in Table 1. The biases are slightly increased from h0 to h10 (Table 1), whereas RMS and SD at h5 and h10 are much smaller than those at h0 (Table 1). In addition, the RMS and its corresponding SD are relatively close to each other at all scales for both the normal and Chi-square distributed data (Table 1). Moreover, SDs in these voxels of ROIs with β3(d0) > 0 are larger than SDs in those voxels of ROI with β3(d0) = 0, since the interior of ROI with β3(d0) = 0 contains more pixels (Figure 4 (c)). Moreover, the SDs at steps h0 and h10 show clear spatial patterns caused by spatial correlations. The RMSs also show some evidence of spatial patterns. The biases, SDs, and RMSs of β3(d0) are smaller in the normal distributed data than in the chi-square distributed data (Table 1), because the signal-to-noise ratios (SNRs) in the normal distributed data are bigger than those SNRs in the chi-square distributed data. Increasing sample size and signal-to-noise ratio decreases the bias, RMS and SD of parameter estimates (Table 1).
Table 1.
Simulation results: Average Bias (×10−2), RMS, SD, and RE of β2(d0) parameters in the five ROIs at 3 different scales (h0, h5, h10), N (0, 1) and χ(3)2 − 3 distributed noisy data, and 2 different sample sizes (n = 60, 80). BIAS denotes the bias of the mean of estimates; RMS denotes the root-mean-square error; SD denotes the mean of the standard deviation estimates; RE denotes the ratio of RMS over SD. For each case, 200 simulated data sets were used.
| χ2(3) − 3 | N(0, 1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 60 | n = 80 | n = 60 | n = 80 | ||||||||||
|
| |||||||||||||
| β2(d0) | h0 | h5 | h10 | h0 | h5 | h10 | h0 | h5 | h10 | h0 | h5 | h10 | |
| 0.0 | BIAS | −0.03 | 0.36 | 0.61 | 0.00 | 0.34 | 0.56 | −0.01 | 0.17 | 0.22 | 0.01 | 0.16 | 0.20 |
| RMS | 0.18 | 0.13 | 0.13 | 0.15 | 0.10 | 0.10 | 0.14 | 0.07 | 0.07 | 0.12 | 0.06 | 0.06 | |
| SD | 0.18 | 0.13 | 0.12 | 0.15 | 0.11 | 0.11 | 0.14 | 0.07 | 0.07 | 0.12 | 0.06 | 0.06 | |
| RE | 1.03 | 1.00 | 1.04 | 1.00 | 0.94 | 0.98 | 0.99 | 0.94 | 1.03 | 1.00 | 0.95 | 1.04 | |
|
| |||||||||||||
| 0.2 | BIAS | 0.72 | 0.37 | 0.38 | 0.15 | −0.35 | −0.39 | −0.04 | −0.55 | −0.66 | 0.10 | −0.48 | −0.61 |
| RMS | 0.19 | 0.14 | 0.13 | 0.16 | 0.11 | 0.11 | 0.14 | 0.07 | 0.07 | 0.12 | 0.06 | 0.06 | |
| SD | 0.18 | 0.14 | 0.13 | 0.16 | 0.12 | 0.11 | 0.14 | 0.08 | 0.07 | 0.12 | 0.07 | 0.06 | |
| RE | 1.02 | 0.99 | 1.03 | 1.00 | 0.96 | 0.99 | 0.99 | 0.96 | 1.04 | 1.00 | 0.97 | 1.06 | |
|
| |||||||||||||
| 0.4 | BIAS | −0.40 | −0.55 | −0.68 | −0.10 | −0.15 | −0.24 | 0.04 | 0.12 | 0.13 | −0.10 | 0.05 | 0.08 |
| RMS | 0.19 | 0.14 | 0.14 | 0.16 | 0.12 | 0.12 | 0.14 | 0.07 | 0.07 | 0.12 | 0.07 | 0.07 | |
| SD | 0.18 | 0.14 | 0.13 | 0.16 | 0.12 | 0.12 | 0.14 | 0.08 | 0.07 | 0.12 | 0.07 | 0.06 | |
| RE | 1.02 | 1.00 | 1.03 | 1.00 | 0.96 | 1.00 | 0.99 | 0.96 | 1.04 | 1.00 | 0.97 | 1.06 | |
|
| |||||||||||||
| 0.6 | BIAS | 0.42 | −1.14 | −1.93 | 0.05 | −1.20 | −1.89 | 0.03 | −0.55 | −0.69 | −0.01 | −0.43 | −0.54 |
| RMS | 0.18 | 0.13 | 0.13 | 0.15 | 0.11 | 0.11 | 0.14 | 0.07 | 0.07 | 0.12 | 0.06 | 0.06 | |
| SD | 0.18 | 0.13 | 0.13 | 0.15 | 0.11 | 0.11 | 0.14 | 0.08 | 0.07 | 0.12 | 0.07 | 0.06 | |
| RE | 1.02 | 1.00 | 1.04 | 1.00 | 0.95 | 0.99 | 0.99 | 0.97 | 1.05 | 1.00 | 0.97 | 1.05 | |
|
| |||||||||||||
| 0.8 | BIAS | −1.04 | −2.95 | −4.09 | −0.13 | −1.71 | −2.70 | −0.11 | −0.82 | −1.03 | −0.03 | −0.59 | −0.77 |
| RMS | 0.19 | 0.15 | 0.15 | 0.16 | 0.12 | 0.12 | 0.14 | 0.08 | 0.07 | 0.12 | 0.07 | 0.07 | |
| SD | 0.19 | 0.15 | 0.14 | 0.16 | 0.13 | 0.12 | 0.14 | 0.08 | 0.07 | 0.12 | 0.07 | 0.06 | |
| RE | 1.02 | 1.00 | 1.03 | 1.00 | 0.96 | 0.99 | 0.99 | 0.94 | 1.01 | 1.00 | 0.95 | 1.02 | |
To assess both Type I and II error rates at the voxel level, we tested the hypotheses H0(d0): βj(d0) = 0 versus H1(d0): βj(d0) ≠ 0 for j = 1, 2, 3 across all d0 ∈
 . We applied the same MASS procedure at scales h0 and h10. The −log10(p) values on some selected slices are shown in the supplementary document. The 200 replications were used to calculate the estimates (ES) and standard errors (SE) of rejection rates at α = 5% significance level. Due to space limit, we only report the results of testing β2(d0) = 0. The other two tests have similar results and are omitted here. For Wβ(d0; h), the Type I rejection rates in ROI with β2(d0) = 0 are relatively accurate for all scenarios, while the statistical power for rejecting the null hypothesis in ROIs with β2(d0) ≠ 0 significantly increases with radius hs and signal-to-noise ratio (Table 2). As expected, increasing n improves the statistical power for detecting β2(d0) ≠ 0.
. We applied the same MASS procedure at scales h0 and h10. The −log10(p) values on some selected slices are shown in the supplementary document. The 200 replications were used to calculate the estimates (ES) and standard errors (SE) of rejection rates at α = 5% significance level. Due to space limit, we only report the results of testing β2(d0) = 0. The other two tests have similar results and are omitted here. For Wβ(d0; h), the Type I rejection rates in ROI with β2(d0) = 0 are relatively accurate for all scenarios, while the statistical power for rejecting the null hypothesis in ROIs with β2(d0) ≠ 0 significantly increases with radius hs and signal-to-noise ratio (Table 2). As expected, increasing n improves the statistical power for detecting β2(d0) ≠ 0.
Table 2.
Simulation Study for Wβ(d0; h): estimates (ES) and standard errors (SE) of rejection rates for pixels inside the five ROIs were reported at 2 different scales (h0, h10), N(0, 1) and χ2(3) − 3 distributed data, and 2 different sample sizes (n = 60, 80) at α = 5%. For each case, 200 simulated data sets were used.
| χ2(3) − 3 | N(0, 1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n = 60 | n = 80 | n = 60 | n = 80 | ||||||
|
| |||||||||
| β2(d0) | s | ES | SE | ES | SE | ES | SE | ES | SE |
| 0.0 | h0 | 0.056 | 0.016 | 0.049 | 0.015 | 0.048 | 0.015 | 0.050 | 0.016 |
| h10 | 0.055 | 0.016 | 0.042 | 0.015 | 0.036 | 0.016 | 0.040 | 0.019 | |
|
| |||||||||
| 0.2 | h0 | 0.210 | 0.043 | 0.245 | 0.039 | 0.282 | 0.033 | 0.370 | 0.035 |
| h10 | 0.358 | 0.126 | 0.413 | 0.139 | 0.777 | 0.107 | 0.870 | 0.081 | |
|
| |||||||||
| 0.4 | h0 | 0.556 | 0.072 | 0.692 | 0.054 | 0.794 | 0.030 | 0.895 | 0.024 |
| h10 | 0.792 | 0.129 | 0.894 | 0.078 | 0.994 | 0.006 | 0.998 | 0.003 | |
|
| |||||||||
| 0.6 | h0 | 0.907 | 0.040 | 0.966 | 0.022 | 0.988 | 0.008 | 0.998 | 0.003 |
| h10 | 0.986 | 0.023 | 0.997 | 0.009 | 1.000 | 0.001 | 1.000 | 0.000 | |
|
| |||||||||
| 0.8 | h0 | 0.978 | 0.016 | 0.997 | 0.004 | 1.000 | 0.001 | 1.000 | 0.000 |
| h10 | 0.997 | 0.006 | 1.000 | 0.001 | 1.000 | 0.000 | 1.000 | 0.000 | |
4 Real Data Analysis
We applied SVCM to the Attention Deficit Hyperactivity Disorder (ADHD) data from the New York University (NYU) site as a part of the ADHD-200 Sample Initiative (http://fcon1000.projects.nitrc.org/indi/adhd200/). ADHD-200 Global Competition is a grassroots initiative event to accelerate the scientific community’s understanding of the neural basis of ADHD through the implementation of open data-sharing and discovery-based science. Attention deficit hyperactivity disorder (ADHD) is one of the most common childhood disorders and can continue through adolescence and adulthood (Polanczyk et al., 2007). Symptoms include difficulty staying focused and paying attention, difficulty controlling behavior, and hyperactivity (over-activity). It affects about 3 to 5 percent of children globally and diagnosed in about 2 to 16 percent of school aged children (Polanczyk et al., 2007). ADHD has three subtypes, namely, predominantly hyperactive-impulsive type, predominantly inattentive type, and combined type.
The NYU data set consists of 174 subjects (99 Normal Controls (NC) and 75 ADHD subjects with combined hyperactive-impulsive). Among them, there are 112 males whose mean age is 11.4 years with standard deviation 7.4 years and 62 females whose mean age is 11.9 years with standard deviation 10 years. Resting-state functional MRIs and T1-weighted MRIs were acquired for each subject. We only use the T1-weighted MRIs here. We processed the T1-weighted MRIs by using a standard image processing pipeline detailed in the supplementary document. Such pipeline consists of AC (anterior commissure) and -PC (posterior commissure) correction, bias field correction, skull-stripping, intensity inhomogeneity correction, cerebellum removal, segmentation, and nonlinear registration. We segmented each brain into three different tissues including grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF). We used the RAVENS maps to quantify the local volumetric group differences for the whole brain and each of the segmented tissue type (GM, WM, and CSF) respectively, using the deformation field that we obtained during registration (Davatzikos et al., 2001). RAVENS methodology is based on a volume-preserving spatial transformation, which ensures that no volumetric information is lost during the process of spatial normalization, since this process changes an individuals brain morphology to conform it to the morphology of the Jacob template (Kabani et al., 1998).
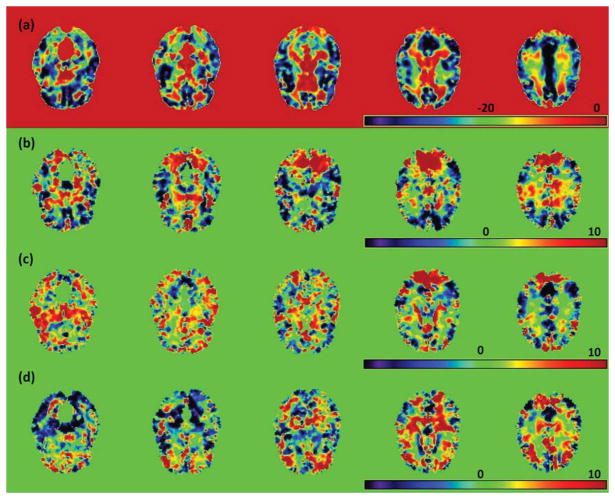
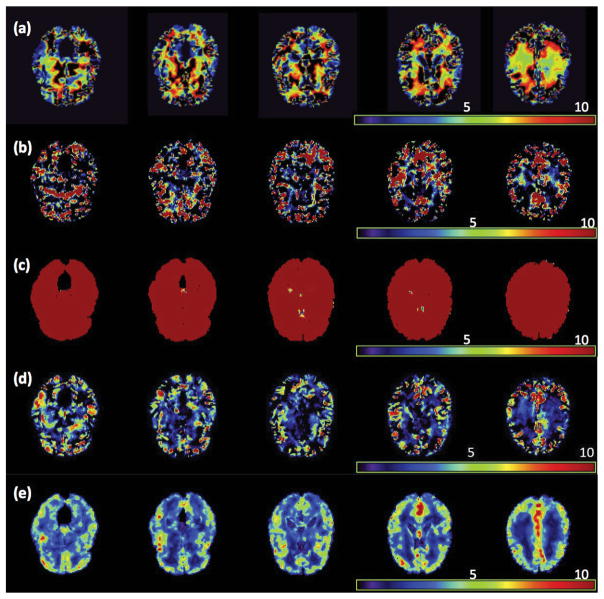
We fitted model (1) to the RAVEN images calculated from the NYU data set. Specifically, we set β(d0) = (β1(d0), …, β8(d0))T and xi = (1, Gi, Ai, Di, WBVi, Ai × Di, Gi × Di, Ai × Gi)T, where Gi, Ai, Di, and WBVi, respectively, represent gender, age, diagnosis (1 for NC and 0 for ADHD), and whole brain volume. We applied the three-stage estimation procedure described in Section 2.2. In MASS, we set hs = 1.1s for s = 1, …, 10. We are interested in assessing the age and diagnosis interaction and the gender and diagnosis interaction. Specifically, we tested H0(d0): β6(d0) = 0 against H1(d0): β6(d0) ≠ 0 for the age × diagnosis interaction across all voxels. Moreover, we also tested H0(d0): β7(d0) = 0 against H1(d0): β7(d0) ≠ 0 for the gender × diagnosis interaction, but we present the associated results in the supplementary document. Furthermore, as shown in the supplementary document, the largest estimated eigenvalue is much larger than all other estimated eigenvalues, which decrease very slowly to zero, and explains 22% of variation in data after accounting for xi. Inspecting Figure 5 reveals that the estimated eigenfunction corresponding to the largest estimated eigenvalue captures the dominant morphometric variation.
Figure 5.
Results from the ADHD 200 data: five selected slices of the four estimated eigenfunctions corresponding to the first four largest eigenvalues of Σ̂η(·, ·): (a) ψ̂1(d); (b) ψ̂2(d); (c) ψ̂3(d); and (d) ψ̂4(d).
As s increases from 0 to 10, MASS shows an advantage in smoothing effective signals within relatively homogeneous ROIs, while preserving the edges of these ROIs (Fig. 6 (a)–(d)). Inspecting Figure 6 (c) and (d) reveals that it is much easier to identify significant ROIs in the −log10(p) images at scale h10, which are much smoother than those at scale h0. To formally detect significant ROIs, we used a cluster-form of threshold of 5% with a minimum voxel clustering value of 50 voxels. We were able to detect 26 significant clusters across the brain. Then, we overlapped these clusters with the 96 predefined ROIs in the Jacob template and were able to detect several predefined ROIs for each cluster. As shown in the supplementary document, we were able to detect several major ROIs, such as the frontal lobes and the right parietal lobe. The anatomical disturbance in the frontal lobes and the right parietal lobe has been consistently revealed in the literature and may produce difficulties with inhibiting prepotent responses and decreased brain activity during inhibitory tasks in children with ADHD (Bush, 2011). These ROIs comprise the main components of the cingulo-frontal-parietal cognitive-attention network. These areas, along with striatum, premotor areas, thalamus and cerebellum have been identified as nodes within parallel networks of attention and cognition (Bush, 2011).
Figure 6.
Results from the ADHD 200 data: five selected slices of (a) β̂6(d0), (b) β̂6(d0; h10), the −log10(p) images for testing H0: β6(d0) = 0 (c) at scale h0 and (d) at scale h10, where β6(d0) is the regression coefficient associated with the age×diagnostic interaction.
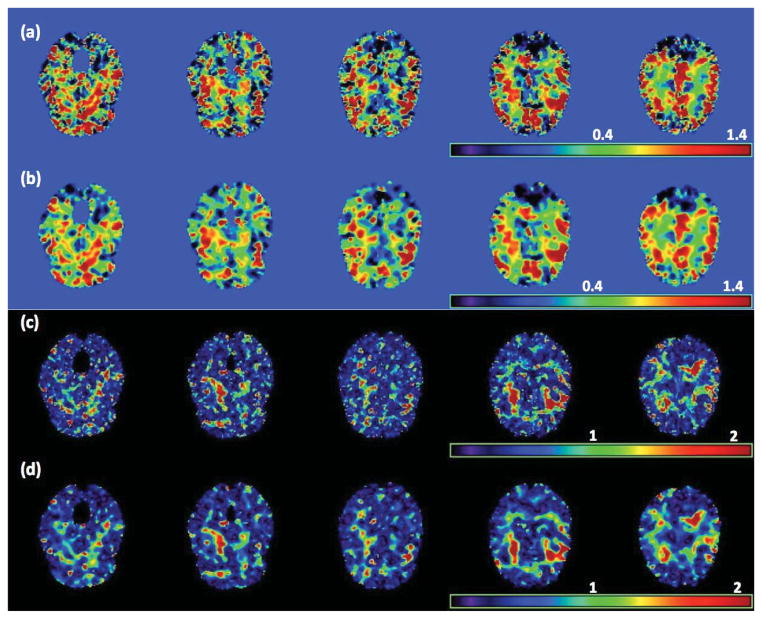
To evaluate the prediction accuracy of SVCM, we randomly selected one subject with ADHD from the NYU data set and predicted his/her RAVENS image by using both model (1) and a standard linear model with normal noise. In both models, we used the same set of covariates, but different covariance structures. Specifically, in the standard linear model, an independent correlation structure was used and the least squares estimates of β(d0) were calculated. For SVCM, the functional principal component analysis model was used and β̂(d0; h10) were calculated. After fitting both models to all subjects except the selected one, we used the fitted models to predict the RAVEN image of the selected subject and then calculated the prediction error based on the difference between the true and predicted RAVEN images. We repeated the prediction procedure 50 times and calculated the mean and standard deviation images of these prediction error images (Figure 7). Inspecting Figure 7 reveals the advantage and accuracy of model (1) over the standard linear model for the ADHD data.
Figure 7.
Results from the ADHD 200 data: The raw RAVENS image for a selected subject with ADHD (a), mean ((b) GLM and (d) SVCM) and standard error ((c) GLM and (e) SVCM) of the errors to predict the RAVENS image in (a), where GLM denotes general linear model.
5 Discussion
This article studies the idea of using SVCM for the spatial and adaptive analysis of neuroimaging data with jump discontinuities, while explicitly modeling spatial dependence in neuroimaging data. We have developed a three-stage estimation procedure to carry out statistical inference under SVCM. MASS integrates three methods including propagation-separation, functional principal component analysis, and jumping surface model for neuroimaging data from multiple subjects. We have developed a fast and accurate estimation method for independently updating each of effect images, while consistently estimating their standard deviation images. Moreover, we have derived the asymptotic properties of the estimated eigenvalues and eigenfunctions and the parameter estimates.
Many issues still merit further research. The basic setup of SVCM can be extended to more complex data structures (e.g., longitudinal, twin and family) and other parametric and semiparametric models. For instance, we may develop a spatial varying coefficient mixed effects model for longitudinal neuroimaging data. It is also feasible to include nonparametric components in SVCM. More research is needed for weakening regularity assumptions and for developing adaptive-neighborhood methods to determine multiscale neighborhoods that adapt to the pattern of imaging data at each voxel. It is also interesting to examine the efficiency of our adaptive estimators obtained from MASS for different kernel functions and coefficient functions. An important issue is that SVCM and other voxel-wise methods do not account for the errors caused by registration method. We may need to explicitly model the measurement errors caused by the registration method, and integrate them with smoothing method and SVCM into a unified framework.
6 Technical Conditions
6.1 Assumptions
Throughout the paper, the following assumptions are needed to facilitate the technical details, although they may not be the weakest conditions. We do not distinguish the differentiation and continuation at the boundary points from those in the interior of
 .
.
Assumption C1. The number of parameters p is finite. Both ND and n increase to infinity such that .
Assumption C2. εi(d) are identical and independent copies of SP(0, Σε) and εi(d) and εi(d′) are independent for d ≠ d′ ∈
 . Moreover, εi(d) are, uniformly in d, sub-Gaussian such that
for all d ∈
. Moreover, εi(d) are, uniformly in d, sub-Gaussian such that
for all d ∈
 and some positive constants Kε and Cε.
and some positive constants Kε and Cε.Assumption C3. The covariate vectors xis are independently and identically distributed with Exi = μx and ||xi||∞ < ∞. Moreover, is invertible. The xi, εi(d), and ηi(d) are mutually independent of each other.
Assumption C4. Each component of {η(d): d ∈
 }, {η(d)η(d′)T: (d, d′) ∈
}, {η(d)η(d′)T: (d, d′) ∈
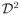 } and {xηT(d): d ∈
} and {xηT(d): d ∈
 } are Donsker classes. Moreover,
} are Donsker classes. Moreover,
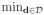 Ση(d, d) > 0 and
for some r1 ∈ (2, ∞), where || · ||2 is the Euclidean norm. All components of Ση(d, d′) have continuous second-order partial derivatives with respect to (d, d′) ∈
Ση(d, d) > 0 and
for some r1 ∈ (2, ∞), where || · ||2 is the Euclidean norm. All components of Ση(d, d′) have continuous second-order partial derivatives with respect to (d, d′) ∈
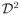 .
.Assumption C5. The grid points
 = {dm, m = 1, …, ND} are independently and identically distributed with density function π(d), which has the bounded support
= {dm, m = 1, …, ND} are independently and identically distributed with density function π(d), which has the bounded support
 . Moreover, π(d) > 0 for all d ∈
. Moreover, π(d) > 0 for all d ∈
 and π(d) has continuous second-order derivative.
and π(d) has continuous second-order derivative.Assumption C6. The kernel functions Kloc(t) and Kst(t) are Lipschitz continuous and symmetric density functions, while Kloc(t) has a compact support [−1, 1]. Moreover, they are continuously decreasing functions of t ≥ 0 such that Kst(0) = Kloc(0) > 0 and limt→∞ Kst(t) = 0.
-
Assumption C7. h converges to zero such that
where c > 0 is a fixed constant and min(q1, q2) > 2.
Assumption C8. There is a positive integer E < ∞ such that λ1 > … > λE ≥ 0.
Assumption C9. For each j, the three assumptions of the jumping surface model hold, each is path-connected, and βj*(d) is a Lipschitz function of d with a common Lipschitz constant Kj > 0 in each such that |βj*(d) − βj*(d′)| ≤ Kj||d − d′||2 for any d, . Moreover,
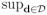 |βj*(d)| < ∞, and max(Kj, Lj) < ∞.
|βj*(d)| < ∞, and max(Kj, Lj) < ∞.- Assumption C10. For piecewise constant βj*(d), and holds uniformly for h0 = 0 < ··· < hS, where Sy =
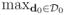 Σy(d0, d0) and u(j)(hs) is the smallest absolute value of all possible jumps at scale hs and given by
Σy(d0, d0) and u(j)(hs) is the smallest absolute value of all possible jumps at scale hs and given by
Assumption C11. For piecewise continuous βj*(d),
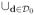 [Pj(d0, hS)c ∩ Ij(d0, δL, δU)] is an empty set and h0 = 0 < h1 < ··· < hS is a sequence of bandwidths such that
, in which limn→∞
Mn = ∞,
and
.
[Pj(d0, hS)c ∩ Ij(d0, δL, δU)] is an empty set and h0 = 0 < h1 < ··· < hS is a sequence of bandwidths such that
, in which limn→∞
Mn = ∞,
and
.
Remark 5
Assumption (C2) is needed to invoke Hoeffding inequality (Buhlmann and van de Geer, 2011; van der Vaar and Wellner, 1996) in order to establish the uniform bound for β̂(d0; hs). In practice, since most neuroimaging data are often bounded, the sub-Gaussian assumption is reasonable. The bound assumption on ||x||∞ in Assumption (C3) is not essential and can be removed if we put a restriction on the tail of the distribution x. Moreover, with some additional efforts, all results are valid even for the case with fixed design predictors. Assumption (C4) avoids smoothness conditions on the sample path η(d), which are commonly assumed in the literature (Hall et al., 2006). The assumption on the moment of is similar to the conditions used in (Li and Hsing, 2010). Assumption (C5) on the stochastic grid points is not essential and can be modified to accommodate the case for fixed grid points with some additional complexities.
Remark 6
The bounded support restriction on Kloc(·) in Assumption (C6) can be weaken to a restriction on the tails of Kloc(·). Assumption (C9) requires smoothness and shape conditions on the image of βj*(d) for each j. For piecewise constant βj*(d), assumption (C10) requires conditions on the amount of changes at jumping points relative to n, ND, and hS. If Kst(t) has a compact support, then Kst(u(j)2/C) = 0 for relatively large u(j)2. In this case, hS can be very large. However, for piecewise continuous βj*(d), assumption (C11) requires the convergence rate of hS and the amount of changes at jumping points.
Supplementary Material
Contributor Information
Hongtu Zhu, Department of Biostatistics and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, hapel Hill, NC 27599, USA.
Jianqing Fan, Department of Oper Res and Fin. Eng, Princeton University, Princeton, NJ 08540.
Linglong Kong, Department of Mathematical and Statistical Sciences, University of Alberta, Edmonton, AB Canada T6G 2G1.
References
- Besag JE. On the statistical analysis of dirty pictures (with discussion) Journal of the Royal Statistical Society, Ser B. 1986;48:259–302. [Google Scholar]
- Buhlmann P, van de Geer S. Statistics for High-Dimensional Data: Methods, Theory and Applications. New York, N.Y: Springer; 2011. [Google Scholar]
- Bush G. Cingulate, frontal and parietal cortical dysfunction in attention-deficit/hyperactivity disorder. Bio Psychiatry. 2011;69:1160–1167. doi: 10.1016/j.biopsych.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Shen J. Image Processing and Analysis: Variational, PDE, Wavelet, and Stochastic Methods. Philadelphia: SIAM; 2005. [Google Scholar]
- Chumbley J, Worsley KJ, Flandin G, Friston KJ. False discovery rate revisited: FDR and topological inference using Gaussian random fields. Neuroimage. 2009;44:62–70. doi: 10.1016/j.neuroimage.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Cressie N, Wikle C. Statistics for Spatio-Temporal Data. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick S. Voxel-based morphome-try using the RAVENS maps: methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Fan J. Local linear regression smoothers and their minimax efficiencies. Ann Statist. 1993;21:196–216. [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modelling and Its Applications. London: Chapman and Hall; 1996. [Google Scholar]
- Fan J, Zhang J. Two-step estimation of functional linear models with applications to longitudinal data. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;62:303–322. [Google Scholar]
- Fan J, Zhang W. Statistical estimation in varying coefficient models. The Annals of Statistics. 1999;27:1491–1518. [Google Scholar]
- Fan J, Zhang W. Statistical methods with varying coefficient models. Stat Interface. 2008;1:179–195. doi: 10.4310/sii.2008.v1.n1.a15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Statistical Parametric Mapping: the Analysis of Functional Brain Images. London: Academic Press; 2007. [Google Scholar]
- Hall P, Müller HG, Wang JL. Properties of principal component methods for functional and longitudinal data analysis. Ann Statist. 2006;34:1493–1517. [Google Scholar]
- Kabani N, MacDonald D, Holmes C, Evans A. A 3D atlas of the human brain. Neuroimage. 1998;7:S717. [Google Scholar]
- Khodadadi A, Asgharian M. Tech rep. McGill University; 2008. Change point problem and regression: an annotated bibliography. http://biostats.bepress.com/cobra/art44. [Google Scholar]
- Lazar NA. The Statistical Analysis of Functional MRI Data. New York: Springer; 2008. [Google Scholar]
- Li SZ. Markov Random Field Modeling in Image Analysis. New York, NY: Springer; 2009. [Google Scholar]
- Li Y, Hsing T. Uniform convergence rates for nonparametric regression and principal component analysis in functional/longitudinal data. The Annals of Statistics. 2010;38:3321–3351. [Google Scholar]
- Li Y, Zhu H, Shen D, Lin W, Gilmore JH, Ibrahim JG. Multiscale adaptive regression models for neuroimaging data. Journal of the Royal Statistical Society: Series B. 2011;73:559–578. doi: 10.1111/j.1467-9868.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Matrix results on the Khatri-Rao and Tracy-Singh products. Linear Algebra Appl. 1999;289:267–277. [Google Scholar]
- Mori S. Principles, methods, and applications of diffusion tensor imaging. In: Toga AW, Mazziotta JC, editors. Brain Mapping: The Methods. 2. Elsevier Science; 2002. pp. 379–397. [Google Scholar]
- Polanczyk G, de Lima M, Horta B, Biederman J, Rohde L. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. The American Journal of Psychiatry. 2007;164:942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- Polzehl J, Spokoiny VG. Adaptive weights smoothing with applications to image restoration. J R Statist Soc B. 2000;62:335–354. [Google Scholar]
- Polzehl J, Spokoiny VG. Propagation-separation approach for local likelihood estimation. Probab Theory Relat Fields. 2006;135:335–362. [Google Scholar]
- Polzehl J, Voss HU, Tabelow K. Structural adaptive segmentation for statistical parametric mapping. NeuroImage. 2010;52:515–523. doi: 10.1016/j.neuroimage.2010.04.241. [DOI] [PubMed] [Google Scholar]
- Qiu P. Image Processing and Jump Regression Analysis. New York: John Wileym & Sons; 2005. [Google Scholar]
- Qiu P. Jump surface estimation, edge detection, and image restoration. Journal of American Statistical Association. 2007;102:745–756. [Google Scholar]
- Ramsay JO, Silverman BW. Functional Data Analysis. New York: Springer-Verlag; 2005. [Google Scholar]
- Scott D. Multivariate Density Estimation: Theory, Practice, and Visualization. New York: John Wiley; 1992. [Google Scholar]
- Spence J, Carmack P, Gunst R, Schucany W, Woodward W, Haley R. Accounting for spatial dependence in the analysis of SPECT brain imaging data. Journal of the American Statistical Association. 2007;102:464–473. [Google Scholar]
- Tabelow K, Polzehl J, Spokoiny V, Voss HU. Diffusion tensor imaging: structural adaptive smoothing. NeuroImage. 2008a;39:1763–1773. doi: 10.1016/j.neuroimage.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Tabelow K, Polzehl J, Ulug AM, Dyke JP, Watts R, Heier LA, Voss HU. Accurate localization of brain activity in presurgical fMRI by structure adaptive smoothing. IEEE Trans Med Imaging. 2008b;27:531–537. doi: 10.1109/TMI.2007.908684. [DOI] [PubMed] [Google Scholar]
- Thompson P, Toga A. A framework for computational anatomy. Computing and Visualization in Science. 2002;5:13–34. [Google Scholar]
- van der Vaar AW, Wellner JA. Weak Convergence and Empirical Processes. Springer-Verlag Inc; 1996. [Google Scholar]
- Wand MP, Jones MC. Kernel Smoothing. London: Chapman and Hall; 1995. [Google Scholar]
- Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. NeuroImage. 2004;23:189–195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Wu CO, Chiang CT, Hoover DR. Asymptotic confidence regions for kernel smoothing of a varying-coefficient model with longitudinal data. J Amer Statist Assoc. 1998;93:1388–1402. [Google Scholar]
- Yue Y, Loh JM, Lindquist MA. Adaptive spatial smoothing of fMRI images. Statistics and its Interface. 2010;3:3–14. [Google Scholar]
- Zipunnikov V, Caffo B, Yousem DM, Davatzikos C, Schwartz BS, Crainiceanu C. Functional principal component model for high-dimensional brain imaging. NeuroImage. 2011;58:772–784. doi: 10.1016/j.neuroimage.2011.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.