Abstract
A nontoxigenic Aspergillus flavus strain, K49, is currently being tested as a biological control agent in corn fields in the Mississippi Delta. However, little is known about the overall genetic diversity of A. flavus from year to year in corn fields and specifically in Mississippi. Our objective was to assess the genetic variability of A. flavus isolates from different seasons, inoculum sources, and years, from a no-till corn field. Of the 175 A. flavus isolates examined, 74 and 97 had the typical norB-cypA type I (1.5 kb) and type II (1.0 kb) deletion patterns, respectively. Variability in the sequence of the omtA gene of the majority of the field isolates (n = 118) was compared to strain K49. High levels of haplotypic diversity (24 omtA haplotypes; Hd = 0.61 ± 0.04) were found. Among the 24 haplotypes, two were predominant, H1 (n = 71), which consists of mostly toxigenic isolates, and H49 (n = 18), which consists of mostly atoxigenic isolates including K49. Toxigenic isolates were prevalent (60%) in this natural population. Nonetheless, about 15% of the population likely shared the same ancestral origin with K49. This study provides valuable information on the diversity of A. flavus. This knowledge can be further used to develop additional biological control strains.
1. Introduction
Corn, Zea mays L., is a major agricultural crop in the state of Mississippi and in the U.S., with an estimated acreage of 770,000 and 91,897,000 acres planted in 2011, respectively [1]. One major food safety concern for corn, peanuts, rice, and edible tree nuts is the contamination with toxins produced by Aspergillus flavus Link [2]. Aspergillus flavus is a soil-borne haploid fungus, known to asexually produce conidia and sclerotia and recently reported to have a sexual stage [3, 4]. The fungus and other Aspergillus spp. in section Flavi can synthesize aflatoxins and cyclopiazonic acid (CPA) [5, 6]. Cyclopiazonic acid is able to inhibit mammalian calcium-ATPase and interfere with Ca2+ level in animal systems [7]. Aflatoxins B1 (AFB1) and B2 (AFB2) are produced by A. flavus, and aflatoxins G1 (AFG1) and G2 (AFG2) in addition to the B aflatoxins are produced by A. parasiticus [8–10]. A. flavus isolates with large deletions of the aflatoxin gene cluster and portions of the subtelomeric region generally result in no aflatoxin and/or CPA production [11].
Aflatoxins can be more problematic for corn grain and residues in the southern U.S. than other regions due to subtropical climate and summer precipitation deficit of the former region [12, 13]. In addition, 70% of corn production uses some form of reduced tillage, therefore increasing the corn residues contaminated with A. flavus left in the field for the next growing season. The U.S. Food and Drug Administration (FDA) regulatory limit for aflatoxin levels in corn feed used in finishing beef cattle is 300 ng/g and for corn used for human consumption is 20 ng/g [14, 15].
A biological control approach to reduce the impact that toxigenic A. flavus strains have on corn production is to use nontoxigenic A. flavus strains to displace the toxigenic strains in the field [16]. These nontoxigenic strains include K49 (NRRL 30797) and the commercially available AF36 (NRRL 18543) and afla-guard (NRRL21882) which are approved by U.S. Environmental Protection Agency (EPA) for use in a variety of crops in the U.S. These strains competitively exclude aflatoxin producers by applying them to soil and plant parts during the vegetative growth stages [17]. International efforts in Argentina [18], Nigeria [19], and China [20] are currently underway to identify prospective biocontrol strains.
Aflatoxins in A. flavus strains are synthesized by a group of approximately 25 genes that form the aflatoxin gene cluster, a region spanning 65–70 kb [10, 25]. Genetic variability occurs in the aflatoxin gene cluster of A. flavus isolates. Partial gene deletions in the norB-cypA gene region result in known deletion patterns I and II [26]. The biocontrol strain AF36 contains all genes in the aflatoxin biosynthesis gene cluster, with a 90% homology to that of aflatoxin producing A. flavus [27]. The basis for its nontoxigenicity is a single nucleotide mutation that results in a stop codon near the beginning of the coding sequence of pksA which encodes a polyketide synthase [28]. Previous studies have confirmed that K49 also contains the whole aflatoxin gene cluster [29]. A recent study reports that the same pksA missense mutation occurs in K49 [30].
A variety of techniques have been used to examine population diversity of toxigenic and nontoxigenic strains present in various crops and geographical locations. They include amplified fragment length polymorphism (AFLP) [31], random amplified fragment polymorphism (RAPD) [32, 33], direct sequencing, intersimple sequence repeats (ISSR) [34], and microsatellite markers [20, 35, 36]. Eight deletion patterns in the aflatoxin gene cluster have been reported for a group of 38 nonaflatoxigenic A. flavus isolates [37]. In a group of soil field isolates from Mississippi corn fields, 60% of A. flavus isolated had the potential to produce aflatoxins and all isolates expressed aflD, aflG, aflP, aflR, and aflS aflatoxin genes [29]. Recently, the presence of mating type genes has been determined (MAT1-1 or MAT1-2) in A. flavus [38]. Moreover, another important target of research is the quantification of A. flavus by real-time PCR that targets the nor-1 gene from the aflatoxin gene cluster [39].
The objective of this research was to assess the temporal genetic variability of A. flavus isolates from a no-tilled corn field in Mississippi using two markers, norB-cypA and omtA genes from the aflatoxin gene cluster. Single nucleotide polymorphism (SNP) in the omtA gene has been shown to have a sufficient discriminatory power in typing strains of A. flavus and closely related A. oryzae [37]. Knowledge of this genetic information will help to better understand the community of isolates in a given corn field with no previous exposure to nontoxigenic strains and to determine if additional local isolates could be developed for biological control programs.
2. Materials and Methods
2.1. Collection of Aspergillus flavus Isolates and Confirmation of Aflatoxin Production
One hundred-seventy five A. flavus isolates were obtained from soil, plant parts (e.g. silk, leaves, cobs, seeds, and tassels), and adult beetles collected randomly from a no-till Bt hybrid corn field in Elizabeth, MS, during the growing (May–October) and overwintering (December–April) seasons of 2006–2009. Soil isolates were obtained as described elsewhere by diluting 10 g of soil into 150 mL of a solution of 2 g of agar/L of water [40]. 100 μL of each sample was transferred to modified dichloronitroaniline rose bengal agar (MDRB) with 3% NaCl and the inoculum was distributed uniformly with a sterile spreader and incubated at 37°C for 5 days [41, 42]. Putative A. flavus isolates were transferred into potato dextrose agar (PDA) plates amended with 0.3%β-cyclodextrin to enhance fluorescence and incubated at 28°C for 5 days in the dark. Aspergillus flavus identification was based on morphological characteristics [43]. Isolates were examined for fluorescence under UV light (365 nm) and subjected to biochemical aflatoxin confirmation with the ELISA Veratox test kit [12, 44].
2.2. DNA Extraction, PCR Amplification, and Sequencing
A single agar plug containing spores was transferred into a sterile 2 mL centrifuge tube containing 1 mL of 0.05% Triton X-100 solution. The tube was vortexed vigorously and 100 μL of the spore suspension was inoculated into 25 mL of potato dextrose broth (PDB) in a 50 mL plastic tube. Tubes were incubated horizontally for 4-5 days at 30°C at 150 rpm.
After visible fungal growth, mycelia was vacuum-dried. Approximately 100–150 mg of wet mycelia was used for DNA extraction using the ZR Fungal/Bacterial DNA kit (Zymo Research, Orange County, CA). DNA extraction followed the manufacturer's instructions in which cells were lysed by bead beating for 5 min with a Disruptor Genie (Zymo Research, Orange County, CA). Eluted DNA was the template for PCR amplification of partial fragments of norB-cypA (300/800 bp) and omtA (594 bp) genes of the aflatoxin gene cluster and ITS (600 bp) gene regions. Platinum Taq DNA Polymerase High Fidelity (Life Technologies, 5791 Van Allen Way, Carlsbad, CA, 92008) was used for the PCR reaction.
Primer pairs were norB-cypA-F 5′-GTGCCCAGCATCTTGGTCCA-3′, norB-cypA-R 5′-AGGACTTGATGATTCCTCGTC-3′ [27], omtA-F 5′-CAGGATATCATTGTGGACGG-3′, omtA-R 5′-CTCCTCTACCAGTGGCTTCG-3′ [36], ITS1 5′-TCCGTAGGTGAACCTGCGG-3′, and ITS4 5′-TCCTCCGCTTATTGATATGC-3′ [20]. ITS sequences of selected samples were used for species confirmation. PCR was conducted following the profile of 2 min at 95°C, followed by 30 cycles of 94°C for 60 s, 45°C for 60 s, and 72°C for 90 s, with a final extension of 72°C for 5 min. Amplicons were separated in 1.5% agarose gels in TBE and photographed using a BioDoc-It Imaging System (UVP Inc., Upland, CA). PCR products for sequencing were gel extracted using Quantum Prep Freeze 'N Squeeze DNA Gel extraction spin columns (Biorad Laboratories, Hercules, CA) and sequenced at the Genomics and Bioinformatics Research Unit of the USDA-ARS in Stoneville, Mississippi.
2.3. Alignment, Population Genetic Analysis, and Population Structure
DNA sequences were aligned with SeqMan Pro in DNASTAR (Madison, WI). omtA haplotypes were determined manually. The program DNAsp version 5.10.01 [45] was used to estimate population genetic parameters: haplotype diversity (Hd), nucleotide diversity (π), mean number of pair wise nucleotide differences (K), theta per site (θ s), and theta per gene (θ g). Levels of gene flow were determined through the effective number of migrants (N m) according to the formula of Nei [24] across years and inoculum source. Parameters that test for population expansion or selection: D + and F + statistics [21], Fu's F's statistic, Strobeck S statistic [22], and Tajima's D [23] statistic, were also calculated with DNAsp.
The program Arlequin version 3.5.1.2 [46] calculated nucleotide differences between years and inoculum sources by estimating F ST values and determined genetic structure with an analysis of molecular variance (AMOVA) with the methodology from Excoffier et al. [47]. Phylogenetic relationships among isolates were established using the Maximum Likelihood (ML) analysis method in MEGA version 5 [48].
3. Results
3.1. Confirmation of Isolates and Deletion Patterns of the norB-cypA Gene Region
All 175 isolates were confirmed with morphological characteristics and also by ITS sequence data (>99.5% homology) to be A. flavus. Conidia of isolates appeared with a characteristic yellowish green to darker green. A total of 105 isolates that were positive for UV fluorescence were also positive for ELISA aflatoxin detection, and 66 were not positive for UV fluorescence and ELISA (Table 1). Amplification of the partial norB-cypA gene region also confirmed that isolates were A. flavus. From a total of 175 isolates, 171 were amplified for the norB-cypA gene region. The norB-cypA F/R primer pair generated a 300 bp and an 800 bp fragment for 74 and 97 A. flavus isolates that correspond to the 1.5 kb (type I) and a 1.0 kb (type II) deletion patterns, respectively (Table 1).
Table 1.
Aspergillus flavus isolates collected in Elizabeth, MS, from 2006 through 2009, UV fluorescence and ELISA aflatoxin detection (+/−), norB-cypA amplicon fragment size, and omtA haplotype frequency.
| Sources | Time period | AF (+) | AF (−) | norB-cypA patterns | omtA haplotypes (frequency) |
|---|---|---|---|---|---|
| Soil | Jan.–Apr. 2006 | 140, 161, 163–172 | 159, 160, 162, 173 | 4b
12c |
H1 (10)d, H6 (1), H14 (1), H17 (1), H49 (2) |
| May–Oct. 2006 | 93–99 | 90–92 | 4b
6c |
H1 (5), H7 (1), H11 (1), H49 (1) | |
| Jan.–Apr. 2007 | 56–58, 60, 76–80, 141, 143–146, 148, 149, 153–155 | 59, 81, 82, 142, 147, 150–152, 156 | 7b
21c |
H1 (15), H16 (1), H19 (1), H49 (6) | |
| May–Oct. 2007 | 101, 105, 107, 110, 115, 123, 138, 139 | 100, 108 | 2b
8c |
H1 (7), H8 (1), H18 (1), H49 (1) | |
| Jan.–Apr. 2008 | 61, 62, 64, 83–88, 157, 158, 175–177, 180 | 63, 65–67, 174, 178, 179 | 6b
14c |
H1 (15), H49 (2) | |
| May–Oct. 2008 | 111, 113, 116, 117, 120, 121, 127, 136 | 132 | 3b
6c |
H1 (4), H5 (1), H15 (1), H21 (1), H49 (1) | |
| Jan.–Apr. 2009 | 55, 73 | 50–54, 68–72, 74, 75, 89 | 8b
7c |
H1 (2), H2 (1), H49 (2) | |
|
| |||||
| Above grounda | May–Oct. 2006 | 124, 128, 134, 137 | 102, 106, 114, 122, 131, 135 | 6b
4c |
H1 (3), H3 (1), H4 (1), H7 (2), H49 (2) |
| May–Oct. 2008 | 104, 109, 112, 118, 125, 126, 129, 133 | 103, 119 | 2b
8c |
H1 (6), H20 (1), H22 (1), H23 (1), H49 (1) | |
|
| |||||
| Plant debris | Jan.–Apr. 2006 | 1–8 | 9–14 | 7b
7c |
H1 (4), H12 (1) |
| Jan.–Apr. 2007 | 19, 22, 24–26, 28, 29 | 16–18, 20, 21, 23, 27, 30 | 15b | H8 (2), H9 (1), H10 (1) | |
| Jan.–Apr. 2008 | 34, 36–38, 40–45 | 31–33, 35, 39 | 11b
4c |
H8 (2), H13 (2) | |
|
| |||||
| Combined | 105 | 66 | 74b, 97c | 118e | |
aAbove ground corresponding to isolates from silk, tassel, ear, and insect.
b300 bp fragment, deletion pattern I.
c800 bp fragment, deletion pattern II.
dNumber of isolates belonging to the indicated haplotype.
eNumber of isolates examined for omtA amplicon.
3.2. Genetic Analysis of omtA Gene Region
A total of 118 A. flavus isolates (67% of n = 175 isolates) were selected for omtA sequence analysis (Table 1). These isolates were selected because their omtA gene fragments were clearly amplified as revealed by gel electrophoresis. The subsequent SNP analysis identified 24 omtA haplotypes. Seventy-one of the 118 isolates belonged to a single haplotype designated as haplotype “1” (H1). This haplotype was observed in 10 of the 12 season/source/year locations. Likewise, haplotype H49, designated “49” because its sequence is exactly the same as K49, was found in 18 isolates in 9 of the 12 season/source/year locations but was not found in plant debris 2006–2008.
Haplotype diversity was high (Hd = 0.61) across sites and ranged from 0.22 to 0.86 (Table 2). The variable number of nucleotide sites (S) totaled 68 from the 597 nucleotide sites analyzed. Of those, singleton variable sites were 31 and parsimony informative sites were 37. Nucleotide diversity (π) was low, ranging between 0.001 and 0.025 with an average of 0.018. Other nucleotide diversity parameters were K = 11.08, θ s = 0.023, and θ g = 13.94 for the total population. The hypothesis of neutral mutation was tested with statistical parameters D +, F +, Fs, Strobeck's S, and Tajima's D. Except for a few parameters, most were not significant at P < 0.05, and hence no assumptions on population growth or selection can be made. Plant debris isolates from the overwintering season of 2007 appear to be most genetically distinct when compared to all other populations. N M in these comparisons was low (≤1.29), which demonstrates low levels of gene flow, and 8 of the 11 F ST comparisons of this group of isolates were significantly different from all other populations tested (Table 3).
Table 2.
omtA gene polymorphism statistics for Aspergillus flavus from a single Mississippi field.
| Season/Source/Year | Sequence and haplotype | Nucleotide diversity | Population expansion | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | S | H | Hd ± sd | π | K | θ s | θ g | D + | F + | Fs | S | D | |
| May–Oct./soil/2006 | 8 | 28 | 4 | 0.64 ± 0.18 | 0.020 | 12.17 | 0.018 | 10.79 | 0.96 | 0.99 | 4.86 | 0.05 | 0.67 |
| May–Oct./soil/2007 | 10 | 26 | 4 | 0.53 ± 0.18 | 0.014 | 8.77 | 0.015 | 9.19 | 1.11 | 0.88 | 5.02 | 0.03 | −0.21 |
| May–Oct./soil/2008 | 8 | 39 | 5 | 0.78 ± 0.15 | 0.023 | 14.25 | 0.025 | 15.42 | 0.15 | 0.02 | 3.32 | 0.16 | −0.40 |
| May–Oct./above ground/2006 | 9 | 28 | 5 | 0.86 ± 0.08 | 0.022 | 13.27 | 0.017 | 10.30 | 1.32 | 1.51 | 3.92 | 0.09 | 1.44 |
| May–Oct./above ground/2008 | 10 | 32 | 5 | 0.66 ± 0.16 | 0.011 | 6.88 | 0.020 | 12.01 | −2.36* | −2.58* | 2.45 | 0.23 | −2.05* |
| Jan.–April/soil/2006 | 15 | 28 | 5 | 0.59 ± 0.14 | 0.015 | 9.27 | 0.014 | 8.80 | 0.92 | 0.84 | 5.34 | 0.02 | 0.22 |
| Jan.–April/soil/2007 | 23 | 26 | 4 | 0.52 ± 0.09 | 0.016 | 9.84 | 0.011 | 7.04 | 1.20 | 1.51 | 11.4 | 0.00 | 1.49 |
| Jan.–April/soil/2008 | 17 | 24 | 2 | 0.22 ± 0.12 | 0.008 | 5.29 | 0.011 | 7.09 | 1.59* | 0.98 | 10.3 | 0.00 | −1.02 |
| Jan.–April/soil/2009 | 5 | 25 | 3 | 0.80 ± 0.16 | 0.024 | 14.80 | 0.020 | 12.00 | 1.73* | 1.86* | 4.74 | 0.09 | 1.73 |
| Jan.–April/plant debris/2006 | 5 | 23 | 2 | 0.40 ± 0.23 | 0.015 | 9.20 | 0.018 | 11.04 | −1.23* | −1.32 | 6.64 | 0.02 | −1.23 |
| Jan.–April/plant debris/2007 | 4 | 23 | 3 | 0.83 ± 0.22 | 0.001 | 1.00 | 0.001 | 1.09 | −0.70 | −0.60 | −0.88 | 0.95 | −0.70 |
| Jan.–April/plant debris/2008 | 4 | 23 | 2 | 0.66 ± 0.20 | 0.025 | 15.33 | 0.021 | 12.54 | 2.28* | 2.38* | 6.76 | 0.03 | 2.28* |
|
| |||||||||||||
| Combined | 118 | 68 | 24 | 0.61 ± 0.04 | 0.018 | 11.08 | 0.023 | 13.94 | −4.07* | −3.13* | 1.83 | 0.20 | −0.65 |
Note: N = number of sequences; S = number of variable sites; H = number of haplotypes; Hd = haplotype diversity ± standard deviation; π = nucleotide diversity; K = mean number of pairwise nucleotide differences; θ s = theta per site; θ g = theta per gene; D +and F + statistics (Fu and Li 1993) [21]; Fs Fu's F's statistic; Strobeck S statistic (Strobeck 1987) [22]; D = Tajima's (1989) [23] statistic; * P < 0.05.
Table 3.
omtA observed F ST values (below) and gene flow (N M) calculated with Nei (1982) [24] for Aspergillus flavus isolates collected from twelve spatiotemporal locations from a single corn field in Mississippi.
| F ST↓ N M→ | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | 1.72 | 2.62 | 11.73 | 10.95 | 29.37 | 0.34 | 10.88 | 3.26 | 4.10 | 1.68 | 2.42 |
| 2 | 0.12 | — | 0.62 | 0.98 | 0.93 | 1.58 | 1.29 | 1.35 | 0.57 | 0.57 | 3.00 | 22.19 |
| 3 | 0.00 | 0.37* | — | 4.51 | 3.78 | 4.77 | 0.11 | 3.11 | 3.86 | 4.74 | 0.58 | 0.65 |
| 4 | −0.05 | 0.29* | −0.00 | — | 87.94 | 42.29 | 0.32 | 15.52 | 9.36 | 18.02 | 1.25 | 1.57 |
| 5 | −0.07 | 0.27* | −0.02 | −0.08 | — | 39.32 | 0.23 | 12.87 | 9.04 | 18.43 | 1.05 | 1.30 |
| 6 | −0.06 | 0.22* | 0.04 | −0.04 | −0.06 | — | 0.60 | 22.67 | 6.48 | 7.77 | 2.22 | 2.99 |
| 7 | 0.54* | 0.17 | 0.77* | 0.65* | 0.67* | 0.57* | — | 0.34 | 0.15 | 0.18 | 0.54 | 0.70 |
| 8 | −0.09 | 0.17 | −0.01 | −0.06 | −0.08 | −0.06 | 0.54* | — | 5.18 | 6.05 | 1.42 | 1.94 |
| 9 | 0.02 | 0.40* | −0.02 | −0.03 | −0.05 | 0.01 | 0.77* | −0.02 | — | 17.39 | 0.63 | 0.74 |
| 10 | 0.03 | 0.44* | 0.00 | −0.03 | −0.05 | 0.01 | 0.79* | −0.00 | −0.05 | — | 0.73 | 0.83 |
| 11 | 0.08 | −0.01 | 0.31 | 0.27* | 0.24* | 0.21 | 0.30 | 0.12 | 0.39* | 0.45* | — | 2.65 |
| 12 | 0.02 | −0.15 | 0.29 | 0.19 | 0.18 | 0.12 | 0.25 | 0.06 | 0.33* | 0.39* | −0.08 | — |
1 = growing season soil 2006; 2 = growing season above ground 2006; 3 = overwintering plant debris 2006; 4 = overwintering soil 2006; 5 = growing season soil 2007; 6 = overwintering soil 2007; 7 = overwintering plant debris 2007; 8 = growing season soil 2008; 9 = growing season above ground 2008; 10 = overwintering soil 2008; 11 = overwintering plant debris 2008; 12 = overwintering soil 2009; *significant at P < 0.05.
3.3. Population Structure
AMOVA partitioned the variation among years, among populations within years and within populations. Variation was mostly explained by within populations (84.92%) (df = 106, F ST = 0.15, P < 0.0009). Variation among populations within years accounted for 20.63% of the variation (df = 8, F SC = 0.19, P < 0.0009) and there was no variation among years (0%) (df = 3, F CT = −0.05, P < 0.56).
3.4. Phylogenetic Analysis
Our ML phylogenetic analysis showed that isolates were not clustered based on the season, inoculum source, or year (Figure 1). For example, isolates 159 to 172 were collected from soil between January and April in 2006 (Table 1), but isolates of 161 and 163–172 are in a clade different from that of isolates 159, 160, and 162. Nonetheless, the 118 isolates formed two major clades based on the aflatoxin producing ability. H1 (isolates 1 to 5) is toxigenic and found to be most distantly related to H49 (H49 to 72). The branch that contains H1, at the upper part of the genealogy tree, includes the group of isolates that were confirmed to be toxigenic. Only 6 isolates within this group were nontoxigenic (isolates 66, 67, 70, 89, 90, 132, and 178) and however closely related to toxigenic isolates by analysis of the omtA gene region. Likewise, there were 7 toxigenic isolates (42, 73, 94, 97, 113, 137, and 148) among the 34 isolates that grouped within the nontoxigenic branch where H49 was positioned. These isolates are closely related to H49 but result in production of aflatoxin.
Figure 1.
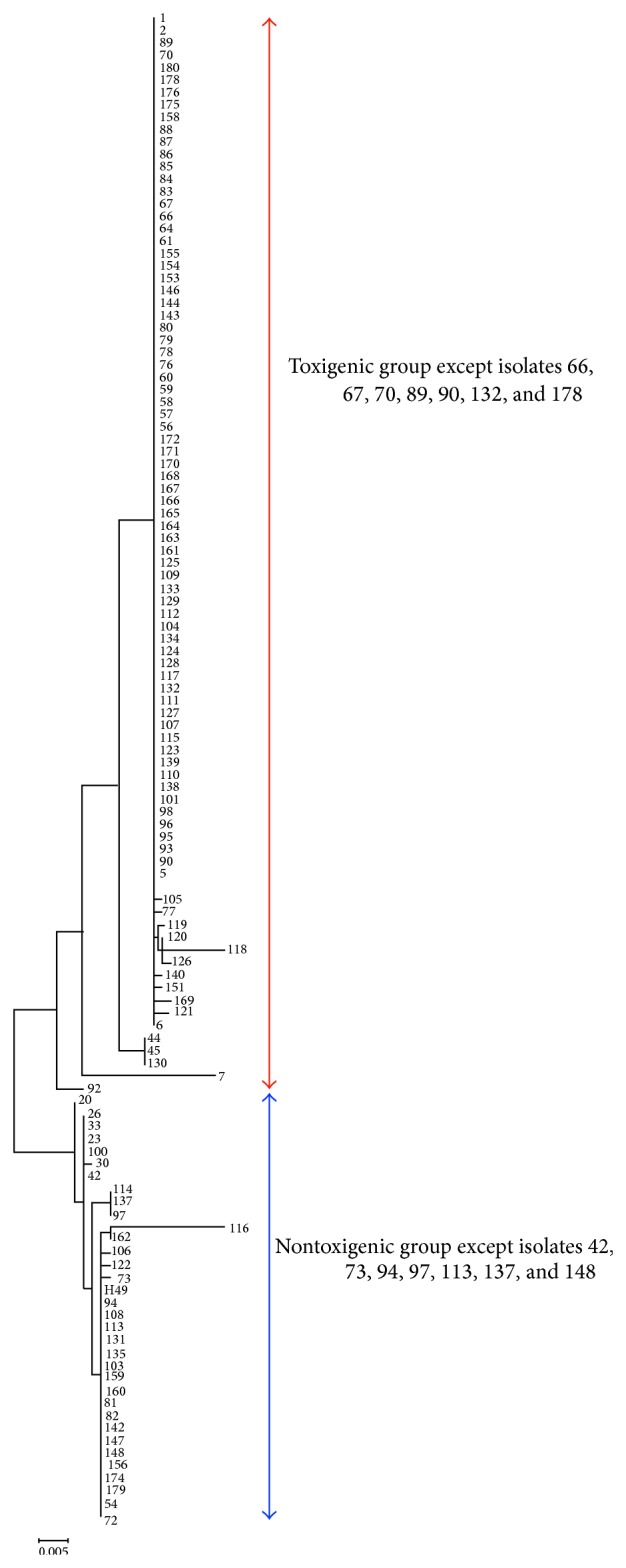
Maximum likelihood analysis of omtA gene sequences for 118 A. flavus collected from a single field in Mississippi 2006–2009. (Scale is 0.005.)
4. Discussion
In this study we assessed the population genetic diversity of A. flavus across years from a single field in Mississippi and determined how this population of isolates is related to the native Mississippi nontoxigenic isolate K49. H49 was present in 10 of the 12 locations, and therefore these isolates could present a shared origin. The other six isolates that were closely related to H49 were nontoxigenic and they may have mutations or deletions in other genes with the aflatoxin gene cluster that result in nontoxigenicity. We found that a high proportion (60%) of the populations are toxigenic isolates of haplotype H1. These H1 toxigenic isolates are present in all years and inoculum sources. In contrast, Abbas et al. [40] observed a higher percent of toxigenic A. flavus isolates in soil than stover and cobs with grain; cobs had the lowest percent of toxigenic A. flavus. We speculate that this no-till field with continuous corn production has supported a buildup of a higher number of H1 toxigenic isolates over time probably in part due to their better competitiveness against A. flavus isolates of other haplotypes.
Aspergillus flavus populations from a corn field community can be diverse due to large amounts of propagule materials [49]. Conidia can be carried through air movement, and sclerotia can remain in the soil for the next growing season. However, we found that, in addition to H1, haplotype H49 is prevalent (15%) in soil and above ground sources but not in plant debris. The biocontrol strain K49 also belongs to the H49 haplotype. Hence, we can assume that H49 isolates from this Mississippi field share an ancestral origin with K49, which was isolated from corn in Mississippi [41]. H49 isolates are present in 10 of the 12 locations examined, which suggests their adaptability and survivability in the field. Other biocontrol candidate strains may be further selected from this particular haplotype. Application of a mixture of highly competitive biocontrol A. flavus strains has become a new paradigm for mitigating preharvest aflatoxin problems in some parts of world, especially in Nigeria [Joseph Atehnkeng personal communication, also see http://www.aflasafe.com/ for details].
Our somewhat limited sample size might explain why a large number of haplotypes were found only once as evidence by the high levels of haplotype diversity. The importance and relative frequency of these single-isolate haplotypes cannot be ascertained unless a larger sample size is taken into consideration. Isolates, detected only once, might have an origin beyond this field and could have been carried in by wind or insects, thereby increasing the genetic diversity of this field. Besides H1 and H49, H7 and H8 are the only other haplotypes found in more than one source. H7 was found in the growing season of 2006 in both soil and above ground inoculum, and H8 was found during the overwintering seasons of 2007 and 2008. Given that there was no structure found across years, variation in the populations cannot be explained as a year effect, but within a year, populations are variable according to their source.
Bayman and Cotty [50] found that 66/105 of A. flavus soil isolates comprised 13 vegetative compatibility groups in a single Arizona cotton field over a 3-year period and one VCG includes 20% of all isolates from the field. Unlike Gashgari et al. [51], who used RAPD analysis, we are able to differentiate toxigenic from nontoxigenic isolates using both analytical methods of UV detection and ELISA and, to a lesser extent, with the partial omtA gene sequence. Others like Midorikawa et al. [33] have been able to characterize A. flavus populations based on host plant. In the context of A. flavus diversity from a single corn field, the omtA marker provided a satisfactory level of resolution. Geiser et al. [52, 53] and Chang et al. [37] included this genetic marker as part of their studies when comparing A. flavus and A. oryzae isolates. Yin et al. [20] also included this genetic marker in the study that examined nontoxigenic peanut A. flavus isolates from a field in China. They found five deletion patterns in 56 toxigenic and nontoxigenic isolates from peanut fields and included 11 genes from the aflatoxin gene cluster in their analysis. The omtA and norB-cypA gene regions can be more commonly used by researchers, as a way to genetically characterize A. flavus isolates. The majority of genes in the aflatoxin gene cluster are highly conserved, as demonstrated by Ehrlich et al. [54] for A. flavus and the related aflatoxin producers of A. nomius and A. parasiticus. The atoxigenic isolates that are clustered with other toxigenic isolates (Figure 1) may have mutations or deletions in genes in the aflatoxin gene cluster, which results in nontoxigenicity [26].
This study provides useful information to the scientific community that studies A. flavus populations. The knowledge of A. flavus genetic diversity can be used to develop additional biocontrol agents native to the local agricultural areas. Further work will determine if these atoxigenic H1 isolates are genetically similar to other genes within the aflatoxin gene cluster, specially the pksA gene that contains the missense mutation that results in nontoxigenicity in the biocontrol strains K49 and AF36.
Acknowledgments
The authors thank Ms. Bobbie Johnson, Ms. Carol Benson, and Mr. Leon Hicks for technical scientific assistance. This research is a contribution to USDA National Program 303.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.USDA-NASS USDA National Agricultural Statistics Service. Grain crops national and state data, http://www.nass.usda.gov/
- 2.Abbas H. K. Aflatoxin and Food Safety. Boca Raton, Fla, USA: CRC Press; 2005. [Google Scholar]
- 3.Amaike S., Keller N. P. Aspergillus flavus . Annual Review of Phytopathology. 2011;49:10.1–10.27. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- 4.Horn B. W., Moore G. G., Carbone I. Sexual reproduction in Aspergillus flavus . Mycologia. 2009;101(3):423–429. doi: 10.3852/09-011. [DOI] [PubMed] [Google Scholar]
- 5.Abbas H. K., Zablotowicz R. M., Horn B. W., Phillips N. A., Johnson B. J., Jin X., Abel C. A. Comparison of major biocontrol strains of non-aflatoxigenic Aspergillus flavus for the reduction of aflatoxins and cyclopiazonic acid in maize. Food Additives and Contaminants. 2011;28(2):198–208. doi: 10.1080/19440049.2010.544680. [DOI] [PubMed] [Google Scholar]
- 6.Abbas H. K., Weaver M. A., Horn B. W., Carbone I., Monacell J. T., Shier W. T. Selection of Aspergillus flavus isolates for biological control of aflatoxins in corn. Toxin Reviews. 2011;30(2-3):59–70. doi: 10.3109/15569543.2011.591539. [DOI] [Google Scholar]
- 7.King E. D., Bassi A. B., Jr., Ross D. C., Druebbisch B. An industry perspective on the use of “atoxigenic” strains of Aspergillus flavus as biological control agents and the significance of cyclopiazonic acid. Toxin Reviews. 2011;30(2-3):33–41. doi: 10.3109/15569543.2011.588818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbas H. K., Wilkinson J. R., Zablotowicz R. M., Accinelli C., Abel C. A., Bruns H. A., Weaver M. A. Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn. Toxin Reviews. 2009;28(2-3):142–153. doi: 10.1080/15569540903081590. [DOI] [Google Scholar]
- 9.Diener U. L., Cole R. J., Sanders T. H., Payne G. A., Lee L. S., Klich M. A. Epidemiology of aflatoxin formation by Aspergillus flavus . Annual Review of Phytopathology. 1987;25:249–270. doi: 10.1146/annurev.py.25.090187.001341. [DOI] [Google Scholar]
- 10.Yu J., Chang P.-K., Ehrlich K. C., Cary J. W., Bhatnagar D., Cleveland T. E., Payne G. A., Linz J. E., Woloshuk C. P., Bennett J. W. Clustered pathway genes in aflatoxin biosynthesis. Applied and Environmental Microbiology. 2004;70(3):1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang P.-K., Horn B. W., Dorner J. W. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus . Fungal Genetics and Biology. 2009;46(2):176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Abbas H. K., Shier W. T., Cartwright R. D. Effet of temperature, rainfall and planting date on aflatoxin and fumonisin contamination in commercial Bt and non-Bt corn hybrids in Arkansas. Phytoprotection. 2007;88(2):41–50. doi: 10.7202/018054ar. [DOI] [Google Scholar]
- 13.Bruns H. A. Controlling aflatoxin and fumonisin in maize by crop management. Journal of Toxicology—Toxin Reviews. 2003;22(2-3):153–173. doi: 10.1081/TXR-120024090. [DOI] [Google Scholar]
- 14.US-FDA (Food and Drug Administration) Action levels for poisonous or deleterious substances in human food and animal feed. http://www.fda.gov/food/guidanceregulation/guidancedocumentsregulatoryinformation/ucm077969.htm.
- 15.van Egmond H. P., Schothorst R. C., Jonker M. A. Regulations relating to mycotoxins in food. Perspectives in a global and European context. Analytical and Bioanalytical Chemistry. 2007;389(1):147–157. doi: 10.1007/s00216-007-1317-9. [DOI] [PubMed] [Google Scholar]
- 16.Bhatnagar D., Cary J. W., Ehrlich K., Yu J., Cleveland T. E. Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development. Mycopathologia. 2006;162(3):155–166. doi: 10.1007/s11046-006-0050-9. [DOI] [PubMed] [Google Scholar]
- 17.Abbas H. K., Zablotowicz R. M., Bruns H. A. Modeling the colonization of maize by toxigenic and non-toxigenic Aspergillus flavus strains: implications for biological control. World Mycotoxin Journal. 2008;1(3):333–340. [Google Scholar]
- 18.Barros G. G., Torres A. M., Rodriguez M. I., Chulze S. N. Genetic diversity within Aspergillus flavus strains isolated from peanut-cropped soils in Argentina. Soil Biology and Biochemistry. 2006;38(1):145–152. doi: 10.1016/j.soilbio.2005.04.028. [DOI] [Google Scholar]
- 19.Donner M., Atehnkeng J., Sikora R. A., Bandyopadhyay R., Cotty P. J. Molecular characterization of atoxigenic strains for biological control of aflatoxins in Nigeria. Food Additives and Contaminants A: Chemistry, Analysis, Control, Exposure and Risk Assessment. 2010;27(5):576–590. doi: 10.1080/19440040903551954. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y., Lou T., Yan L., Michailides T. J., Ma Z. Molecular characterization of toxigenic and atoxigenic Aspergillus flavus isolates, collected from peanut fields in China. Journal of Applied Microbiology. 2009;107(6):1857–1865. doi: 10.1111/j.1365-2672.2009.04356.x. [DOI] [PubMed] [Google Scholar]
- 21.Fu Y.-X., Li W.-H. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strobeck C. Average number of nucleotide differences in a sample from a single subpopulation: a test for population subdivision. Genetics. 1987;117:149–153. doi: 10.1093/genetics/117.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei M. Evolution of human races at the gene level. In: Bonne-Tamir B., Cohen T., Goodman R. M., editors. Human Genetics Part A: The Unfolding Genome. New York, NY, USA: Alan R. Liss; 1982. pp. 167–181. [Google Scholar]
- 25.Payne G. A., Yu J., Nierman W. C., Machida M., Bhatnagar D., Cleveland T. E., Dean R. A. A first glance into the genome sequence of Aspergillus flavus . In: Osmani S. A., Goldman G. H., editors. The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods. Boca Raton, Fla, USA: CRC Press; 2008. pp. 15–23. [Google Scholar]
- 26.Chang P.-K., Ehrlich K. C., Hua S.-S. T. Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. International Journal of Food Microbiology. 2006;108(2):172–177. doi: 10.1016/j.ijfoodmicro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Das M. K., Ehrlich K. C., Cotty P. J. Use of pyrosequencing to quantify incidence of a specific Aspergillus flavus strain within complex fungal communities associated with commercial cotton crops. Phytopathology. 2008;98(3):282–288. doi: 10.1094/PHYTO-98-3-0282. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich K. C., Cotty P. J. An isolate of Aspergillus flavus used to reduce aflatoxin contamination in cottonseed has a defective polyketide synthase gene. Applied Microbiology and Biotechnology. 2004;65(4):473–478. doi: 10.1007/s00253-004-1670-y. [DOI] [PubMed] [Google Scholar]
- 29.Accinelli C., Abbas H. K., Zablotowicz R. M., Wilkinson J. R. Aspergillus flavus aflatoxin occurrence and expression of aflatoxin biosynthesis genes in soil. Canadian Journal of Microbiology. 2008;54(5):371–379. doi: 10.1139/W08-018. [DOI] [PubMed] [Google Scholar]
- 30.Chang P.-K., Abbas H. K., Weaver M. A., Ehrlich K. C., Scharfenstein L. L., Cotty P. J. Identification of genetic defects in the atoxigenic biocontrol strain Aspergillus flavus K49 reveals the presence of a competitive recombinant group in field populations. International Journal of Food Microbiology. 2012;154(3):192–196. doi: 10.1016/j.ijfoodmicro.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Wicklow D. T., McAlpin C. E., Platis C. E. Characterization of the Aspergillus flavus population within an Illinois maize field. Mycological Research. 1998;102(3):263–268. doi: 10.1017/S0953756297004851. [DOI] [Google Scholar]
- 32.Lourenço A., Durigon E. L., Zanotto P., Madeira J. E. G. C., De Almeida A. P., Correa B. Genetic diversity of environmental Aspergillus flavus strains in the state of São Paulo, Brazil by random amplified polymorphic DNA. Memorias do Instituto Oswaldo Cruz. 2007;102(6):687–692. doi: 10.1590/S0074-02762007000600005. [DOI] [PubMed] [Google Scholar]
- 33.Midorikawa G. E. O., Pinheiro M. R. R., Vidigal B. S., Arruda M. C., Costa F. F., Pappas G. J., Jr., Ribeiro S. G., Freire F., Miller R. N. G. Characterization of Aspergillus flavus strains from Brazilian Brazil nuts and cashew by RAPD and ribosomal DNA analysis. Letters in Applied Microbiology. 2008;47(1):12–18. doi: 10.1111/j.1472-765X.2008.02377.x. [DOI] [PubMed] [Google Scholar]
- 34.Batista P. P., Santos J. F., Oliveira N. T., Pires A. P. D., Motta C. M. S., Luna-Alves Lima E. A. Genetic characterization of Brazilian strains of Aspergillus flavus using DNA markers. Genetics and Molecular Research. 2008;7(3):706–717. doi: 10.4238/vol7-3gmr422. [DOI] [PubMed] [Google Scholar]
- 35.Grubisha L. C., Cotty P. J. Twenty-four microsatellite markers for the aflatoxin-producing fungus Aspergillus flavus . Molecular Ecology Resources. 2009;9(1):264–267. doi: 10.1111/j.1755-0998.2008.02378.x. [DOI] [PubMed] [Google Scholar]
- 36.Hatti A. D., Taware S. D., Taware A. S., Pangrikar P. P., Chavan A. M., Mukadam D. S. Genetic diversity of toxigenic and non-toxigenic Aspergillus flavus strains using ISSR markers. International Journal of Current Research. 2010;5:61–66. [Google Scholar]
- 37.Chang P.-K., Horn B. W., Dorner J. W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genetics and Biology. 2005;42(11):914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Prado J. H., Moore G. G., Horn B. W., Carbone I. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus . Fungal Genetics and Biology. 2008;45(9):1292–1299. doi: 10.1016/j.fgb.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Passone M. A., Rosso L. C., Ciancio A., Etcheverry M. Detection and quantification of Aspergillus section Flavi spp. in stored peanuts by real-time PCR of nor-1 gene, and effects of storage conditions on aflatoxin production. International Journal of Food Microbiology. 2010;138(3):276–281. doi: 10.1016/j.ijfoodmicro.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Abbas H. K., Accinelli C., Zablotowicz R. M., Abel C. A., Bruns H. A., Dong Y., Shier W. T. Dynamics of mycotoxin and Aspergillus flavus levels in aging Bt and non-Bt corn residues under Mississippi no-till conditions. Journal of Agricultural and Food Chemistry. 2008;56(16):7578–7585. doi: 10.1021/jf801771a. [DOI] [PubMed] [Google Scholar]
- 41.Abbas H. K., Zablotowicz R. M., Weaver M. A., Horn B. W., Xie W., Shier W. T. Comparison of cultural and analytical methods for determination of aflatoxin production by Mississippi Delta Aspergillus isolates. Canadian Journal of Microbiology. 2004;50(3):193–199. doi: 10.1139/w04-006. [DOI] [PubMed] [Google Scholar]
- 42.Horn B. W., Dorner J. W. Soil populations of Aspergillus species from section Flavi along a transect through peanut-growing regions of the United States. Mycologia. 1998;90(5):767–776. doi: 10.2307/3761317. [DOI] [Google Scholar]
- 43.Klich M. A. Identification of Common Aspergillus Species. Utrecht, The Netherlands: Centraalbureau voor Schimmelcultures; 2002. [Google Scholar]
- 44.Abbas H. K., Williams W. P., Windham G. L., Pringle H. C., III, Xie W., Shier W. T. Aflatoxin and fumonisin contamination of commercial corn (Zea mays) hybrids in Mississippi. Journal of Agricultural and Food Chemistry. 2002;50(18):5246–5254. doi: 10.1021/jf020266k. [DOI] [PubMed] [Google Scholar]
- 45.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 46.Excoffier L., Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 47.Excoffier L., Smouse P. E., Quattro J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zablotowicz R. M., Abbas H. K., Locke M. A. Population ecology of Aspergillus flavus associated with Mississippi Delta soils. Food Additives and Contaminants. 2007;24(10):1102–1108. doi: 10.1080/02652030701546198. [DOI] [PubMed] [Google Scholar]
- 50.Bayman P., Cotty P. J. Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Canadian Journal of Botany. 1991;69(8):1707–1711. doi: 10.1139/b91-216. [DOI] [Google Scholar]
- 51.Gashgari R. M., Shebany Y. M., Gherbawy Y. A. Molecular characterization of mycobiota and aflatoxin contamination of retail wheat flours from jeddah markets. Foodborne Pathogens and Disease. 2010;7(9):1047–1054. doi: 10.1089/fpd.2009.0506. [DOI] [PubMed] [Google Scholar]
- 52.Geiser D. M., Pitt J. I., Taylor J. W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus . Proceedings of the National Academy of Sciences of the United States of America. 1998;95(1):388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geiser D. M., Dorner J. W., Horn B. W., Taylor J. W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae . Fungal Genetics and Biology. 2000;31(3):169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- 54.Ehrlich K. C., Yu J., Cotty P. J. Aflatoxin biosynthesis gene clusters and flanking regions. Journal of Applied Microbiology. 2005;99(3):518–527. doi: 10.1111/j.1365-2672.2005.02637.x. [DOI] [PubMed] [Google Scholar]


