Fig. 1.
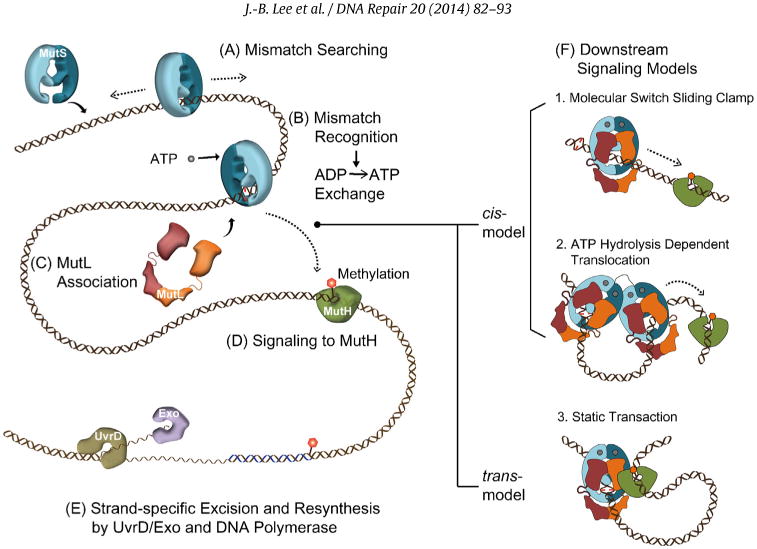
The mechanism of mismatch repair. (A) Mismatch searching performed by MutS homologs (MSH) occurs on duplex DNA. (B) Mismatch binding by an MSH provokes the exchange of bound ADP for ATP (ADP → ATP exchange). (C) The MutL homolog (MLH/PMS) associates with the MSH to transmit the mismatch binding signal. (D) In bacteria signal, the first downstream signaling component appears to be MutH that is responsible for introducing a strand scission on the newly synthesized unmethylated DNA strand. Methylation is controlled by the DNA adenine methylase (Dam) at GATC sequences. (E) Excision of the newly replicated strand is controlled by the UvrD helicase along with one of four exonucleases (see text) in concert with the MSH-MLH/PMS complex. Resynthesis of the single-stranded gap is performed by the replicative DNA polymerase. (F) Models for mismatch signal transmission: (1) Molecular Switch Sliding Clamp where ATP-bound MSH along with MLH/PMS moves by 1-dimensional thermal diffusion to downstream mismatch repair (MMR) machinery, (2) ATP-Hydrolysis Dependent Translocation where the MSH and MLH/PMS form a multimeric complex that translocates to the downstream machinery using the energy of ATP hydrolysis forming a bidirectional loop structure, and (3) Static Transactivation where an MSH-MLH/PMS complex is formed at the mismatch that then associates with downstream MMR machinery by 3D collision forming a static loop structure.
