Abstract
Clinical isolates of Klebsiella pneumoniae are divided into three phylogroups and differ in their virulence factor contents. The aim of this study was to determine an association between phylogroup, virulence factors and mortality following bloodstream infection (BSI) caused by Klebsiella pneumoniae. Isolates from all adult patients with BSI caused by K. pneumoniae admitted to Karolinska University Hospital, Solna between 2007 and 2009 (n = 139) were included in the study. Phylogenetic analysis was performed based on multilocus sequence typing (MLST) data. Testing for mucoid phenotype, multiplex PCR determining serotypes K1, K2, K5, K20, K54 and K57, and testing for virulence factors connected to more severe disease in previous studies, was also performed. Data was retrieved from medical records including age, sex, comorbidity, central and urinary catheters, time to adequate treatment, hospital-acquired infection, and mortality, to identify risk factors. The primary end-point was 30- day mortality. The three K. pneumoniae phylogroups were represented: KpI (n = 96), KpII (corresponding to K. quasipneumoniae, n = 9) and KpIII (corresponding to K. variicola, n = 34). Phylogroups were not significantly different in baseline characteristics. Overall, the 30-day mortality was 24/139 (17.3%). Isolates belonging to KpIII were associated with the highest 30-day mortality (10/34 cases, 29.4%), whereas KpI isolates were associated with mortality in 13/96 cases (13.5%). This difference was significant both in univariate statistical analysis (P = 0.037) and in multivariate analysis adjusting for age and comorbidity (OR 3.03 (95% CI: 1.10–8.36). Only three of the isolates causing mortality within 30 days belonged to any of the virulent serotypes (K54, n = 1), had a mucoid phenotype (n = 1) and/or contained virulence genes (wcaG n = 1 and wcaG/allS n = 1). In conclusion, the results indicate higher mortality among patients infected with isolates belonging to K. variicola. The increased mortality could not be related to any known virulence factors, including virulent capsular types or mucoid phenotype.
Introduction
Klebsiella pneumoniae is a gram-negative pathogen causing a wide spectrum of both hospital- and community-acquired infections such as urinary tract infection, pneumonia, intraabdominal infection, bloodstream infection (BSI), meningitis and pyogenic liver abscess (PLA) [1]–[6]. The mortality in invasive infection is high, ranging between 17.5 and 23% [7]–[10].
Recently, K. pneumoniae has emerged as an increasingly resistant pathogen; it has shown an unprecedented ability to express several intrinsic and acquired mechanisms making the species frequently multidrug-resistant (MDR) to clinically important antimicrobial classes [11]. The clonal dissemination of resistant strains has been the focus of significant attention because of the lack of effective treatment options [12],[13]. Therefore, the dramatic increase in antibiotic-resistant outbreaks faced in several hospital and geographic locations has a high potential of causing mortality and morbidity [14]–[18].
K. pneumoniae displays a large spectrum of virulence factors associated with the infective potential of community-acquired-isolates [19]–[22]. The bacterial capsule is important for its virulence, with 78 capsular types identified [23],[24]. Among them K1, K2, K5, K20, K54, K57 were associated with invasive disease [6],[19],[21],[25]. Other potential pathogenic factors are the lipopolysaccharide [26],[27], iron scavenging systems, fimbrial and non-fimbrial adhesion factors [28],[29], hypermucoviscosity [30],[31], the plasmid-borne rmpA [32] (regulator of mucoid phenotype A), wcaG; encoding the capsular fucose synthesis, which may enhance the antiphagocytic activities [33], and allS; encoding the activator of allantoin regulon [34].
Klebsiella pneumoniae isolates display metabolic versatility, enabling bacteria of this genus to thrive in a variety of environmental niches [1],[20],[21],[35].
Phylogenetic analysis of clinical, carriage and environmental isolates classically identified as K. pneumoniae demonstrated the existence of three main phylogenetic lineages (phylogroups) called KpI, KpII and KpIII as initially demonstrated based on the sequence analysis of gyrA and parC genes [36],[37] and later by their association with specific families of chromosomal β-lactamase genes [38],[39]. From a taxonomical standpoint, Klebsiella pneumoniae comprises three subspecies [1],[36],[40],[41]: K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozaenae and K. pneumoniae subsp. rhinoscleromatis. The two latter subspecies are rarely encountered and are associated to specific diseases (rhinoscleroma and ozena, respectively). From a genetic viewpoint, these two subspecies represent homogeneous genotypic groups (MLST clonal complexes; CC) that are nested within the main phylogroup of K. pneumoniae, named KpI [36]. Subsequent taxonomic work has proposed the names Klebsiella variicola, for phylogroup KpIII [42] and K. quasipneumoniae for phylogroup KpII [43]. As a consequence, the name K. pneumoniae should now be used only for strains that belong to phylogroup KpI (K. pneumoniae sensu stricto).
It is difficult to distinguish K. pneumoniae from K. quasipneumoniae and K. variicola by biochemical tests [42],[44]. Until now, K. variicola and K. quasipneumoniae strains have therefore been generally misidentified as K. pneumoniae in clinical microbiology laboratories. Currently these three species can be more reliably differentiated by genotyping methods [36],[37],[45]. The introduction of mass spectrometry might offer a possibility of rectifying this problem in the future. As a result, the clinical importance of K. variicola and K. quasipneumoniae is currently underestimated, and there are few studies that have reported these species from clinical samples [37],[46]. K. variicola isolates were frequently isolated from various plants [42]. Previous investigations have shown that approximately 20% of human isolates thought to be K. pneumoniae are in fact K. variicola/KpIII or K. quasipneumoniae/KpII [37],[42].
A number of studies have shown association between pathogenicity of K. pneumoniae and virulence genes, serotypes, sequence types and mucoid phenotypes [21],[47]–[49]. The purpose of this study was to characterize clinical invasive isolates of K. pneumoniae sensu lato (i.e., in the classical sense and including K. variicola, K. quasipneumoniae and K. pneumoniae sensu stricto) from the Stockholm area and to analyze their population structure. A specific focus was to identify the main phylogroups. Further, our aims were to analyze their clonal diversity; to describe the association of virulence factors and serotypes with phylogroups; to study the association between phylogroups, bacterial traits and severity of disease, with primary endpoint being 30-day mortality.
Materials and Methods
Patients and bacterial isolates
All adult (≥18 years old) patients admitted to Karolinska University Hospital, Solna, Sweden, between 2007 and 2009, with growth of K. pneumoniae either in blood (n = 137) or cerebrospinal fluid (CSF) (n = 1) or both (n = 2) were included in this retrospective cohort study. Isolates and patients were detected by searches in the clinical microbiology laboratory information system. Species identification was done with API 20E system (bioMérieux, Marcy l’Etoile, France) or VITEK2 (bioMérieux). Antimicrobial susceptibility testing was performed with the disk diffusion method on Isosensitest agar (Oxoid, Basingstoke, UK) interpreted according to the guidelines of the Swedish Reference Group for Antibiotics (SRGA) [50]. Genotypic analysis of isolates producing extended-spectrum β-lactamases and/or carbapenemases was performed with Check-MDR (Checkpoints, Wageningen, The Netherlands). One isolate was excluded due to non-typability with MLST, hence the total amount of included isolates were 139. One blood or CSF isolate from each patient was used for the subsequent analyses.
Capsule typing and virulence gene detection
Detection of serotypes and virulence genes was performed by PCR [51],[52]. We sought for six major serotypes that have been reported to be strongly associated with community-acquired invasive disease: K1, K2, K5, K20, K54, and K57. The virulence genes allS, rmpA, and wcaG were also investigated as described previously [21],[53]. Hypermucoviscous phenotype was determined using the string test, i.e. by pulling colonies with an inoculation loop, using a threshold of 5 mm [6].
MLST
MLST was carried out as previously described [21],[54]. Updated details of protocol and primers for amplification and sequencing are given on the web site (http://bigsdb.web.pasteur.fr). Templates were sequenced on both strands with the published primers using the BigDye Terminator Ready Reaction Mix v3.1. Nucleotide sequences were determined by ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). New allelic variants were repeated and confirmed in triplicate. Alleles and profiles were determined by direct comparison with the K. pneumoniae MLST database. The alignment of sequences was performed with the Mega v5 [55]. Neighbor-joining trees were drawn for concatenate sequence of the seven MLST loci for the total population. To identify phylogenetic groups, three reference strains (group KpI: K. pneumoniae ATCC13883: ST3; group KpII: SB59: ST1118; and group KpIII: Kp342: ST146) were included in phylogenetic analyses [36]. The phylogenetic structure was also assessed by drawing separately a Neighbor-joining tree for each allele. Phylogenetic groupings were identified also by NeighborNet analysis based on the concatenated sequences using SplitsTree v4 [56].
DnaSP v5 [57] was used to calculate the nucleotide diversity of each gene for each phylogroup and for the whole population. To provide a graphical representation of the populations, a minimal spanning tree (MST) [58] was produced with BioNumerics v7.0 (Applied Maths, Sint Maartens-Latem, Belgium). Clonal complexes were defined as groups of isolates sharing six loci of their allelic profiles with at least one other member of the group, using the program eBURST v3 [59].
Clinical parameters
The medical records were retrieved for the 139 cases, studying mortality, patient risk factors, hospital- versus community-acquired disease and antibiotic treatment. Charlson comorbidity index were constructed to assess comorbidity [60]. Infection was classified as polymicrobial if at least one more different species was recovered from specimens drawn within 24 h from the K. pneumoniae bloodstream infection. Skin contaminants (i.e., Corynebacterium spp., Bacillus spp., Propionibacterium spp., and coagulase-negative staphylococci [CoNS]) had to be present in at least two blood cultures, while for other species presence in one blood culture was sufficient. [61] Infections were defined as hospital-acquired if the sample was taken>48 h after admittance to hospital or if the patient had been admitted to hospital within the previous 30 days.
Statistical analysis
Fisher’s test and Chi-squared test were used as appropriate for categorical variables. The Mann Whitney test was used to compare continuous variables. For all tests a two-sided p-value <0.05 was considered significant. Odds Ratio (OR) and 95% Confidence Intervals (CI) were calculated using logistic regression in the STATA 12.0 software. The multivariate analysis was restricted by the relatively few outcomes (mortality in analyzed groups, n = 23). Since our primary interest was to investigate the effect of phylogeny on mortality risk we decided to build a first model by including age and Charlson comorbidity index to adjust for host factors (these have previously been correlated to severe outcome in patients with severe bacterial disease). Both were adjusted for as continuous variables. In a second model we further included polymicrobial infection (present/absent) and gender. To avoid adjusting for factors that could be on the causal pathway between phylogroup and mortality, we did not adjust for disease manifestation (severity of disease and location). Time to adequate antibiotic therapy could be regarded as being on the causal pathway since it is likely influenced by disease manifestation. However, since it is often included in multivariate models of bacterial strains/clones and mortality we included time to adequate therapy as a continuous variable in a third model for comparison.
Ethical considerations
Ethical approval for the study was obtained from the Karolinska Institutet Regional Ethics Committee of Stockholm (recordal 2009/1985-31/4). The committee approved that no written or verbal consent had to be given by the study subjects, as the study only pertained to extracting limited clinical data from patient charts. Patient records were anonymized and de-identified prior to analysis.
Results
Phylogenetic diversity and analysis of MLST data
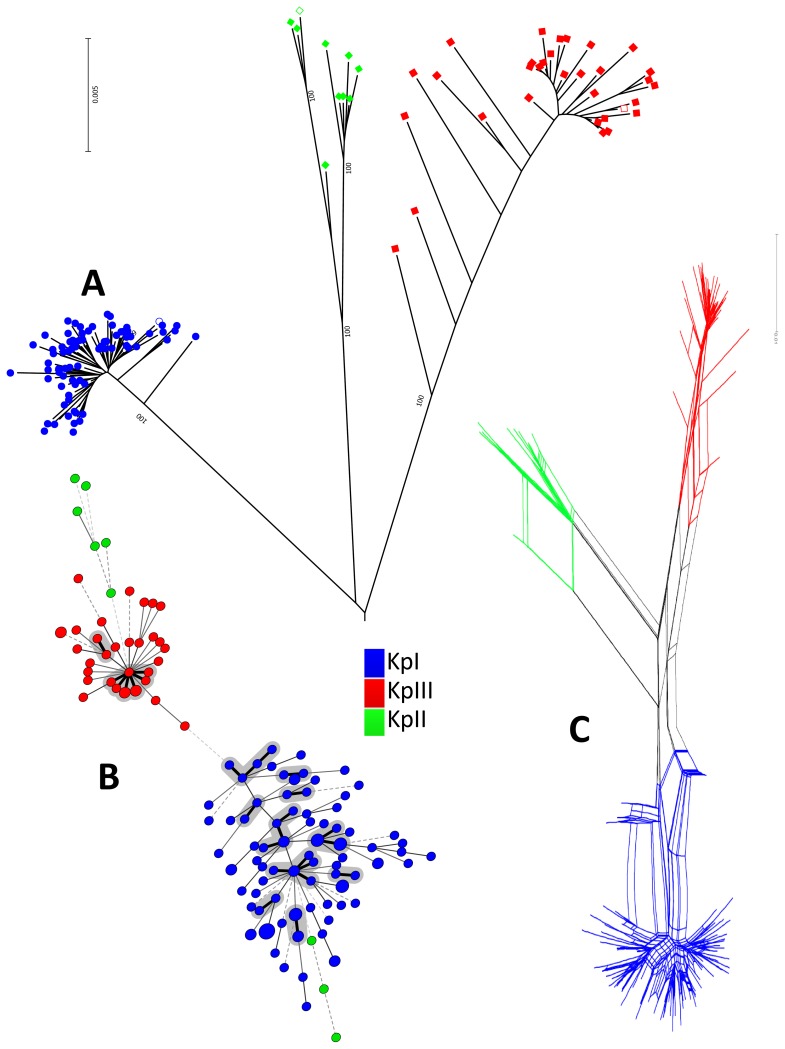
The phylogenetic analysis of the concatenated sequences of the 7 MLST loci (Fig. 1A, Fig. S1) clearly revealed a well-defined structure, with three phylogroups that were each supported by maximal bootstrap values. The obtained phylogeny corroborates previous findings [36],[37],[42]. The three clades in the figures correspond to phylogroups KpI, KpII and KpIII (Fig. 1A, B, Fig. S1). KpI was the largest phylogroup consisting of 96 isolates, followed by KpIII (Klebsiella variicola) consisting of 34 isolates, and KpII (K. quasipneumoniae) consisting of 9 isolates. As previously noted [36], KpI and KpII were branched together while KpIII was branched in an external position (Fig. 1A). The mean genetic distance between clusters was 4.0% (KpI-KpII); 4.4% (KpI-KpIII) and 4.5% (KpII-KpIII). Furthermore the three phylogroups were confirmed by conducting a Neighbor-joining tree analysis with each of the individual alleles (data not shown). The NeighborNet graph (Fig. 1C) also demonstrated a clear demarcation among the three phylogroups and suggested a high level of recombination within each of the groups as well as among them.
Figure 1. Phylogenetic representation of all isolates.
A: Radial phylogenetic tree of 139 isolates based on the concatenated sequences of seven MLST loci performed using the Neighbor-joining method based on a Jukes-Cantor distance matrix. Main bootstrap values obtained are highlighted on at the main node of the phylogeny. The tree was rooted using the nucleotide sequences of the seven genes of E. coli and considered as an out-group. Each phylogroup was clearly separated from others. Colors of isolates symbols are specific of each phylogroups. Blue color corresponds to the KpI phylogroup, green corresponds to KpII, and red corresponds to KpIII. Empty symbols correspond to the references strains specific for each phylogroups. B: Minimal Spanning Tree (MST) analysis of Klebsiella pneumoniae strains based on MLST allelic profiles. Each circle corresponds to an ST. The area of each circle corresponds to the number of isolates. The relationships between strains are indicated by the connections between the isolates and the lengths of the branches linking them. Black lines connecting pairs of STs indicate that they differ in one allele (thick lines), two and three alleles (thin), or four to seven alleles (dashed). Colors of isolates symbols are specific to each phylogroup: blue - KpI, green - KpII and red - KpIII. Grey zones surround STs differing in one allele forming a clonal complex. C: NeighborNet graph based on seven concatenated housekeeping genes, illustrating the recombination within and among phylogroups. Colors surrounding each zone are specific of each phylogroup. Blue color corresponds to the KpI phylogroup, green corresponds to KpII, and red corresponds to KpIII.
The alignment of the seven housekeeping genes revealed the presence of minor events of insertion/deletion (INDEL) only in the tonB locus. The polymorphism parameters were assessed in the whole population, and in the three phylogroups (Table 1). Considering all isolates, the polymorphic sites ranged from 5.1% (gapA) to 19.6% (tonB), whereas the concatenate sequence revealed 10.8% polymorphic sites (Table 1). The average nucleotide diversity π was 2.1% for all the population. However, the internal level of π within the three phylogroups was much lower and the diversity within KpI (0.37%) and KpIII (0.63%) was lower than for KpII (1.3%). A ratio of Ka/Ks <1 was observed, as typical for housekeeping genes evolving predominantly under purifying selection.
Table 1. Nucleotide polymorphism within the seven housekeeping genes used for MLST.
| Alleles | n | Sites | NetSites | S | Hap | Hd | π | θ | Ka/Ks |
| gapA | 139 | 450 | 450 | 23 | 18 | 0.692 | 0.00819 | 0.009683 | 0.09948677 |
| infB | 139 | 318 | 318 | 28 | 19 | 0.756 | 0.015221 | 0.017127 | 0 |
| Mdh | 139 | 477 | 477 | 59 | 26 | 0.805 | 0.027967 | 0.023598 | 0.03242491 |
| Pgi | 139 | 432 | 432 | 54 | 28 | 0.693 | 0.02405 | 0.023955 | 0.01440774 |
| phoE | 139 | 420 | 420 | 41 | 41 | 0.939 | 0.014013 | 0.020316 | 0.00640582 |
| rpoB | 139 | 501 | 501 | 41 | 27 | 0.815 | 0.014476 | 0.015582 | 0.02302211 |
| tonB | 139 | 438 | 402 | 79 | 47 | 0.952 | 0.046675 | 0.040646 | 0.06934406 |
| Concatenate, Total 139 strains | 139 | 3036 | 3000 | 325 | 115 | 0.996 | 0.021386 | 0.021363 | 0.03443384 |
| Concatenate, KpI | 96 | 3024 | 3006 | 115 | 75 | 0.993 | 0.003715 | 0.007448 | 0.07781457 |
| Concatenate, KpII | 9 | 3018 | 3012 | 105 | 9 | 1 | 0.013206 | 0.012949 | 0.02047211 |
| Concatenate, KpIII (K. variicola) | 34 | 3018 | 3006 | 155 | 31 | 0.995 | 0.006379 | 0.012936 | 0.01251564 |
N = number of STs or samples subjected for analysis. Sites = length of sequences after alignments. Net Sites = length of sequences subjected for analysis (the gaps or missing data are rejected from analysis). S = polymorphic sites. Hap = haplotypes. Hd = haplotypic diversity. π = nucleotide diversity. θ = average number of nucleotide differences per site. Ka/Ks = ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks).
The population analysis based on the MLST data showed an extensive haplotypic diversity, with 116 distinct STs. The eBURST and MST analyses (Fig. 1B) were both used to depict the clonal relatedness among sequence types (STs). The 139 isolates were subdivided into 12 clonal complexes comprising 48 isolates (Fig. 1B, Table 2). The remaining 91 isolates were divided into 79 singletons (STs that differed by two or more alleles from any other ST).
Table 2. eBURST analysis of the MLST data: illustration of clonal complexes based on STs and their allelic profiles.
| Clonal Complexes | STs | Allelic profiles |
| CC347 | 347 | 16, 24, 21, 27, 47, 22, 67 |
| 596 | 16, 24, 21, 27, 41, 22, 67 | |
| 595 | 16, 24, 21, 27, 55, 22, 67 | |
| 619 | 64, 24, 21, 27, 47, 22, 67 | |
| 197 | 16, 28, 21, 27, 47, 22, 67 | |
| 642 | 16, 24, 21, 24, 47, 22, 67 | |
| 597 | 16, 24, 21, 27, 47, 22, 105 | |
| CC253 | 253 | 2, 1, 1, 1, 9, 1, 13 |
| 704 | 2, 1, 1, 1, 9, 1, 20 | |
| 163 | 2, 1, 1, 1, 9, 1, 12 | |
| 588 | 2, 1, 1, 1, 9, 1, 23 | |
| 618 | 2, 1, 1, 1, 116, 1, 13 | |
| CC25 | 25 | 2, 1, 1, 1, 10, 4, 13 |
| 65 | 2, 1, 2, 1, 10, 4, 13 | |
| 280 | 2, 1, 2, 1, 10, 4, 46 | |
| 304 | 2, 1, 1, 1, 10, 4, 90 | |
| CC268 | 268 | 2, 1, 2, 1, 7, 1, 81 |
| 703 | 16, 1, 2, 1, 7, 1, 7 | |
| 701 | 2, 1, 2, 2, 7, 1, 81 | |
| 36 | 2, 1, 2, 1, 7, 1, 7 | |
| CC17 | 17 | 2, 1, 1, 1, 4, 4, 4 |
| 20 | 2, 3, 1, 1, 4, 4, 4 | |
| 336 | 2, 1, 1, 1, 72, 4, 4 | |
| CCI | 504 | 2, 1, 1, 1, 3, 3, 18 |
| 540 | 2, 1, 71, 1, 3, 3, 18 | |
| CCII | 35 | 2, 1, 2, 1, 10, 1, 19 |
| 693 | 2, 1, 2, 1, 10, 3, 19 | |
| CCIII | 357 | 16, 24, 21, 27, 54, 22, 45 |
| 599 | 16, 24, 21, 27, 111, 22, 45 | |
| CCIV | 541 | 4, 1, 2, 1, 1, 1, 4 |
| 584 | 4, 1, 2, 1, 1, 7, 4 | |
| CCV | 15 | 1, 1, 1, 1, 1, 1, 1 |
| 14 | 1, 6, 1, 1, 1, 1, 1 | |
| CCVI | 13 | 2, 3, 1, 1, 10, 1, 19 |
| 591 | 2, 3, 1, 1, 7, 1, 19 | |
| CCVII | 593 | 2, 1, 2, 1, 9, 1, 16 |
| 429 | 2, 1, 2, 1, 9, 1, 116 |
CCI to CCVII are an arbitrary designations of clonal complexes found in the present study, each complex formed only by two STs.
Capsule types and virulence factors content of phylogroups
Among the 139 isolates (Fig. S2B, Table S1) only 18 strains (12.9%) were serotypable for the investigated capsular serotypes; K1 (1.4%; 2/139), K2 (5.0%; 7/139), K20 (1.4%; 2/139), K54 (2.2%; 3/139); K57 (2.9%; 4/139). K2 was the most frequently occurring serotype and K5 was not detected in any of the isolates. Of the 18 capsular types found in this study, 16 belonged to KpI and 2 to KpIII.
Regarding virulence genes (Fig. S2C, Table S1), rmpA was positive for 5 strains (3.6%; 5/139; ST25, 380, 592, 593 and 701), wcaG was positive for 12 strains (8.6%; 12/139; ST14, 29, 29, 581, 599, 604, 605, 616, 637, 695, 699 and 700) and allS was positive for 6 strains (4.3%; 6/139; ST104, 604, 605, 613, 620 and 702). A mucoid phenotype (Fig. S2D, Table S1) was detected in 8 isolates (5.8%; 8/139; ST20, 111, 268, 280, 380, 504, 542 and 592) out of which two were also positive for rmpA. With the exception of ST380, which was previously associated with rmpA [47], there were no other observed correlations between virulence genes and STs or clonal complexes. The 21 isolates with virulence genes were distributed as follows: KpI (n = 13), KpIII (n = 5) and KpII (n = 3).
Association of clinical features with phylogroups
Among the 139 infectious episodes, 24 events were associated with a fatal outcome within 30 days translating to a total 30-day mortality of 17.3% (Table S1). Isolates belonging to phylogroup KpIII had the highest 30-day mortality (29.4%, 10/34 patients; Fig. S2A, Table S1). In phylogroup KpI and KpII the 30-day mortality was 13.5% (13/96) and 11.1% (1/9) (Fig. S2A, Table S1), respectively. In further comparisons of the association between phylogroup and mortality phylogroup KpII was excluded due to the small amount of patients. There was a significant difference in 30-day mortality between KpIII and KpI in crude analyses (OR 2.66 (95% CI: 1.04–6.82, p = 0.037) (Table 3). Increasing Charlson comorbidity index was strongly associated with mortality (p = 0.005). Metastatic cancer, which attributes 6 points to the Charlson comorbidity index, was also associated with mortality (p = 0.001). After adjustment for age and Charlson comorbidity index, KpIII remained associated with increased mortality (OR: 3.03 (95% CI: 1.10–8.36). Furthermore, after including age, Charlson comorbidity index, presence of polymicrobial infection and gender, as well as inclusion of time to adequate antibiotic therapy, KpIII was associated with increased mortality [OR: 3.11 (95% CI: 1.10–8.80) and 3.23 (95% CI: 1.12–9.27) respectively]. In a final analysis we excluded 7 patients that were treated at the intensive care unit at the time of onset of the BSI, which did not alter our results (KpIII adjusted OR: 3.53 (95% CI: 1.20–10.39).
Table 3. Comparison between patients with 30-day mortality and survivors.
| Dead within 30 days (n = 23) No. (%) | Survivors>30 days (n = 107) No. (%) | Dead vs. Survivors Significance P<0.05 | |
| Clinical characteristics | |||
| Median age, years | 73 | 68 | 0.35 |
| Male sex | 14 (60.9) | 70 (65.4) | 0.68 |
| Charlson index (median) | 5 | 2 | 0.0003 |
| Charlson index 0–1 | 1 (4.4) | 17 (15.9) | 0.005 |
| Charlson index 2–3 | 8 (34.8) | 62 (57.9) | |
| Charlson index 4–5 | 3 (13.0) | 12 (11.2) | |
| Charlson index>5 | 11 (47.8) | 16 (15.0) | |
| Diabetes | 2 (8.7) | 20 (18.7) | 0.25 |
| Heart disease | 8 (34.8) | 28 (26.2) | 0.40 |
| Pulmonary disease | 7 (30.4) | 15 (14.0) | 0.070 |
| Kidney disease | 4 (17.4) | 18 (16.8) | 1.00 |
| Liver disease | 6 (26.1) | 16 (15.0) | 0.22 |
| CNS-disease | 5 (21.7) | 17 (15.9) | 0.54 |
| Intestinal disease | 3 (13.0) | 13 (12.1) | 1.00 |
| Malignancy, all | 15 (62.5) | 60 (56.1) | 0.42 |
| Hematological | 3 (13.0) | 21 (19.6) | 0.57 |
| Urogenital | 5 (21.7) | 18 (16.8) | 0.56 |
| Colorectal | 1 (4.3) | 10 (9.3) | 0.69 |
| Pulmonary | 3 (13.0) | 3 (2.8) | 0.068 |
| Bile/liver/pancreas | 1 (4.3) | 5 (4.7) | 1.00 |
| Miscellaneous | 2 (8.7) | 3 (2.8) | 0.21 |
| Metastasized | 10 (43.5) | 12 (11.2) | 0.001 |
| Previous organ transplant | 1 (4.3) | 2 (1.9) | 0.45 |
| Neutropenia | 2 (8.7) | 19 (17.8) | 0.37 |
| Hospital-acquired infection | 11 (47.8) | 54 (50.5) | 0.82 |
| Urinary catheter | 9 (39.1) | 41 (38.3) | 0.94 |
| Central catheter | 7 (30.4) | 33 (30.8) | 0.97 |
| Polymicrobial | 8 (34.8) | 24 (22.4) | 0.21 |
| E coli | 5 (21.7) | 9 (8.4) | 0.074 |
| Source of infection | |||
| Urinary | 7 (30.4) | 47 | 0.23 |
| Respiratory tract | 3 (13.0) | 5 (4.7) | 0.15 |
| Bile/liver | 1 (4.3) | 10 | 0.69 |
| Gastrointestinal tract | 3 (13.0) | 6 (5.6) | 0.20 |
| Miscellaneous | 3 (13.0) | 10 | 0.59 |
| Unknown | 6 (26.1) | 29 (27.1) | 0.92 |
| Time to adequate antimicrobial therapy | |||
| 0–1 h | 2 (8.7) | 12 (11.2) | 0.98 |
| 1–2 h | 6 (26.1) | 21 (19.6) | |
| 2–4 h | 6 (26.1) | 28 (26.2) | |
| 4–24 h | 6 (26.1) | 33 (30.8) | |
| 24–48 | 2 (8.7) | 9 (8.4) | |
| >48 h | 1 (4.4) | 4 (3.7) | |
| Bacterial characteristics | |||
| KpIII* | 10 (43.5) | 24 (22.4) | 0.037 |
| Mucoid phenotype | 1 (4.3) | 7 (6.5) | 1.00 |
| Serotypability | 1 (4.3) | 17 (15.9) | 0.20 |
| Virulence genes | 1 (4.3) | 17 (15.9) | 0.20 |
| Any virulence factor | 2 (8.7) | 29 (27.1) | 0.060 |
| ESBL | 1 (4.3) | 4 (3.7) | 1.00 |
*Compared to KpI where the prevalence was 56.5% (13/23) in patients who died and. 77.6% (83/107) in patients who survived.
Among the 24 events associated with 30-day mortality only one isolate was serotypable (K54), and strikingly few (three isolates) featured virulence genes. Almost all isolates belonged to new sequence types, with the exception of ST15 (forming a clonal complex with ST14, which is prevalent among carbapenemase-producers) [53],[62],[63]; ST37 (previously described in bloodstream infection) [54]; ST337 (previously described in bloodstream infection in KPC-producing isolate, Colombia) [64],[65]; ST359 (previously described in bloodstream infection, colistin-resistant, South Korea) [66] and ST280 (previously described in bovine mastitis) (http://bigsdb.web.pasteur.fr). Table 4 summarizes patient characteristics. A majority of the patients (62.6%) were male, and the median age was 70 years. Comorbidity measured in Charlson comorbidity index ranged from 0 to 9. Most of the patients, 76/139 (54.7%) had an index of 2–3, 18 (24.5%), 15 (10.8%) and 30 (21.6%) had Charlson comorbidity index of 0–1, 4–5 and>5 respectively. The most common disease was having a malignancy with a prevalence of 81/139 (58.3%). Hematological malignancy was most common (n = 26) followed by urogenital malignancies and colorectal cancer (n = 25 and n = 11, respectively). 68/139 (48.9%) of the infectious episodes were considered hospital-acquired. Polymicrobial infections occurred in almost one fourth of the patients (34/139; 24.5%). The most frequent co-pathogen in the polymicrobial infections, observed in 15/139 (10.8%) patients, was Escherichia coli followed by Staphylococcus aureus n = 5 (3.6%) and Enterococcus spp, n = 5 (3.6%) (E. faecalis n = 2, E. faecium n = 2 and E. casseliflavus n = 1) respectively. There were no significant differences between the phylogenetic groups concerning patient characteristics (Table 4).
Table 4. Clinical characteristics of patients with invasive infection caused by K. pneumoniae/variicola .
| All (n = 139) No. (%) | KpI (n = 96) No. (%) | KpIII (n = 34) No. (%) | KpI vs. KpIII Significance P<0.05 | |
| Age, median, years | 70 | 65.1 | 68 | 0.62 |
| Male sex | 87 (62.6) | 58 (60.4) | 26 (76.5) | 0.093 |
| Charlson index, median | 2 (0–9) | 2 (0–9) | 3 (1–7) | 0.37 |
| Charlson index 0–1 | 18 (12.9) | 17 (17.7) | 1 (2.9) | 0.07 |
| Charlson index 2–3 | 76 (54.7) | 47 (49.0) | 23 (67.7) | |
| Charlson index 4–5 | 15 (10.8) | 10 (10.4) | 5 (14.7) | |
| Charlson index>5 | 30 (21.6) | 22 (22.9) | 5 (14.7) | |
| Diabetes | 24 (17.3) | 15 (15.6) | 7 (20.6) | 0.51 |
| Heart disease | 38 (27.3) | 25 (26.0) | 11 (32.4) | 0.48 |
| Pulmonary disease | 25 (18.0) | 14 (14.6) | 8 (23.5) | 0.23 |
| Kidney disease | 25 (18.0) | 17 (17.7) | 5 (14.7) | 0.69 |
| Liver disease | 22 (15.8) | 16 (16.7) | 6 (17.6) | 0.90 |
| CNS-disease | 24 (17.3) | 18 (18.8) | 5 (14.7) | 0.60 |
| Intestinal disease | 17 (12.2) | 13 (13.5) | 6 (17.6) | 0.56 |
| Malignancy, all | 81 (58.3) | 51 (53.1) | 21 61.8) | 0.38 |
| Hematologic | 26 (18.7) | 16 (16.7) | 8 (23.5) | 0.38 |
| Urogenital | 25 (18.0) | 18 (18.8) | 5 (14.7) | 0.60 |
| Colorectal | 11 (7.9) | 7 (7.3) | 4 (11.8) | 0.48 |
| Pulmonary | 7 (5.0) | 4 (4.2) | 3 (8.8) | 0.38 |
| Bile/liver/pancreas | 6 (4.3) | 6 (6.3) | 0 | 0.34 |
| Miscellaneous | 6 (4.3) | 3 (3.1) | 2 (5.9) | 0.61 |
| Metastasized | 26 (18.7) | 18 (18.8) | 5 (14.7) | 0.60 |
| Prev. organ transplant | 4 (2.9) | 3 (3.1) | 0 (0) | 0.57 |
| Neutropenia | 24 (17.3) | 14 (14.6) | 7 (20.6) | 0.42 |
| Hospital-acquired infection | 68 (48.9) | 50 (52.1) | 14 (41.2) | 0.27 |
| Urinary catheter | 54 (38.8) | 40 (41.7) | 10 (29.4) | 0.21 |
| Central catheter | 43 (30.9) | 28 (29.2) | 12 (35.3) | 0.51 |
| Source of infection | ||||
| Urinary | 59 (42.4) | 42 (43.8) | 12 (35.3) | 0.39 |
| Respiratory tract | 8 (5.8) | 6 (6.3) | 2 (5.9) | 1.00 |
| Bile/liver | 11 (7.9) | 9 (9.4) | 2 (5.9) | 0.73 |
| Gastrointestinal tract | 9 (6.5) | 4 (4.2) | 5 (14.7) | 0.052 |
| Miscellaneous | 14 (10.1) | 11 (11.5) | 2 (5.9) | 0.51 |
| Unknown | 38 (27.3) | 26 (27.1) | 11 (32.4) | 0.63 |
| Polymicrobial (all) | 34 (24.5) | 21 (21.9) | 11 (32.4) | 0.22 |
| E. coli | 15 (10.8) | 9 (9.4) | 5 (14.7) | 0.52 |
| S. aureus | 5 (3.6) | 3 (3.1) | 1 (2.9) | 1.00 |
| Enterococcus spp | 5 (3.6) | 2 (2.1) | 2 (5.9) | 0.28 |
Antibiotic treatment and antimicrobial resistance
The most common antibiotic treatment, in 58.3% (81/139) of the patients, was cephalosporins (cefuroxime, cefotaxime or ceftazidime) (Table S1). Twenty-six patients (18.7%) received piperacillin-tazobactam as empiric treatment, while 13.7% (19/139) and 6.5% (9/139) started with a carbapenem (imipenem or meropenem) and ciprofloxacin, respectively. Aminoglycosides (gentamicin, amikacin) were added to the first dose of empiric treatment in 23/139 cases (16.5%) and in 4 of these cases this component was the only active against the K. pneumoniae isolate.
Within 4 hours, adequate antibiotic treatment was given to 77/139 patients (55.4%), 44/139 received adequate treatment within 2 hours (31.7%), and 15/139 (10.8%) within 1 hour (Table S1). After 24 hours 121/139 patients (87.1%) had received appropriate treatment, and after 48 hours most patients, 134/139 (96.4%) had received adequate antibiotic treatment.
Overall there was a low level of acquired antibiotic resistance (Table 5). The resistance levels against antimicrobials with relevant activities were as follows: trimethoprim/sulfametoxazole 10.8%, ciprofloxacin 8.6%, piperacillin/tazobactam 5.8%, gentamicin 3.6%, ceftazidime 4.3% and cefotaxime 3.6%. The resistance against trimethoprim/sulfametoxazole was significantly higher in KpI than compared to KpIII (P = 0.014). A total of 15.1% of the isolates were resistant to one or several of these antibiotic classes, 18.8% in KpI and 5.9% in KpIII (P = 0.078). These finding are similar to previous reports [37],[46] in which K. variicola isolates have been found less resistant than KpI and KpII for several classes of antimicrobial agents.
Table 5. No of isolates with non-susceptibility (intermediate and resistant) to antimicrobials.
| Antimicrobial | All (139) | KpI (n = 96) | KpIII (n = 34) | KpI vs. KpIII Significance P<0.05 |
| Ciprofloxacin | 12 (8.6) | 10 (10.4) | 2 (5.9) | 0.73 |
| Trimethoprim/sulfametoxazole | 15 (10.8) | 15 (15.6) | 0 | 0.014 |
| Cefotaxime | 5 (3.6) | 5 (5.2) | 0 | 0.33 |
| Ceftazidime | 6 (4.3) | 5 (5.2) | 0 | 0.33 |
| Gentamicin | 5 (3.6) | 5 (5.2) | 0 | 0.33 |
| Piperacillin/tazobactam | 8 (5.8) | 8 (8.3) | 0 | 0.11 |
| Meropenem | 1 (0.7) | 1 (1.0) | 0 | 1.00 |
| Multidrug-resistance (I or R ≥3 classes of antibiotics) | 5 (3.6) | 5 (5.2) | 0 | 0.33 |
| ESBL* (including CPE) | 5 (3.6) | 5 (5.2) | 0 | 0.33 |
| CPE (ESBLCARBA)** | 1 (0.7) | 1 (1.0) | 0 | 1.00 |
| Total isolates with resistance to any group above | 21 (15.1) | 18 (18.8) | 2 (5.9) | 0.074 |
CPE = carbapenemase-producing Enterobacteriaceae.
*Two isolates CTX-M-1, one SHV-ESBL, one CTX-M-1+SHV-ESBL in genotypic analysis.
**VIM, CTX-M-1, CMY-2 in genotypic analysis.
Five of 139 isolates (3.6%) were considered multidrug-resistant (resistant to ≥three of the following antibiotic classes; trimethoprim/sulfametoxazole, fluoroquinolones, piperacillin/tazobactam, cephalosporins, aminoglycosides or carbapenems) [67]. These isolates all produced extended-spectrum β-lactamases (ESBL); two isolates CTX-M group 1 (ST15), one isolate CTX-M- group1 and an SHV-ESBL (ST340) and one isolate an SHV-ESBL (ST17). The fifth isolate (ST383) co-produced a CTX-M group 1 enzyme, CMY II, and the carbapenemase VIM. The patient had previously been hospitalized in Greece [68]. All ESBL-producing isolates belonged to KpI. Among the 4 patients with carbapenem susceptible ESBL-producing isolates, two received adequate empiric antibiotic treatment within 4 hours (carbapenems), in the other two cases there was a delay between 24 and 72 hours until adequate treatment was received (no death occurred). The patient with BSI caused by a VIM-producing strain received adequate antibiotic treatment, colistin, rifampicin and meropenem, after 4 days and had a fatal outcome. However this patient also had significant comorbidities requiring ICU-care, respirator and dialysis before the onset of the bloodstream infection.
Discussion
To our knowledge this study is the first merging phylogroup classification, clinical perspective, virulence factors and sequence types on an inpatient consecutive material of bloodstream infections caused by Klebsiella pneumoniae (sensu lato) during a long time period and in a well-defined geographical area. The population structure of K. pneumoniae has been suggested to consist of three phylogroups [36] but this has not been thoroughly explored in specific geographical locations and with respect to clinical outcome and patient characteristics. According to our findings, the Swedish K. pneumoniae population comprises the three main phylogroups (Fig. 1A, B, C). KpI was the most frequent group consisting 96 isolates (69%), followed by KpIII consisting of 34 isolates (24%) and KpII consisting of 9 isolates (6%). Previously [37], 420 isolates from 26 European hospitals were classified as follows: 345 isolates (82.1%) identified as KpI, 29 (6.9%) as KpII and 46 (11%) as KpIII. Hence, the population in this study was comparable, in terms of phylogroup relative frequencies, to a Europe-wide sampling [37]. Phylogroups KpII and KpIII were given species status as K. quasipneumoniae and K. variicola [42],[43]. These observations confirm that K. pneumoniae, as defined biochemically and identified in clinical microbiology laboratories, must be regarded as a complex of three species. These three species appear to have wide geographic distribution and to be represented, among isolates initially identified as K. pneumoniae, in similar relative order of frequency in distinct geographical locations [46],[52].
Phylogroup KpI has been shown to be overrepresented among clinical K. pneumoniae isolates in previous studies [36],[46]. Contrary to previous studies, phylogroup KpIII (K. variicola) isolates were relatively more common in this study. Recently Seki et al. [69] described a case of fatal bloodstream infection caused by K. variicola. Initially, based on the classic and automated identification methods, the isolate was incorrectly identified as K. pneumoniae, but reclassified as K. variicola according to data derived from next-generation sequencing. As it is difficult to distinguish K. variicola from K. pneumoniae by classical methods used in clinical laboratories for species determination, K. variicola has been underreported in the literature. K. variicola was initially shown to be adonitol negative unlike K. pneumoniae [36],[42], but this test alone cannot reliably identify K. variicola [43],[46]. Utilization of 5-keto-D-gluconate was recently found to be specific of K. variicola and may represent a useful feature for identification [43]. Also, the usefulness of mass spectrometry for species identification of K. variicola remains to be determined.
Here we addressed for the first time to our knowledge, the possible presence of a link between phylogroup and bacterial traits (virulence factor, serotypes) or severity of disease. The overall 30-day mortality was 17.3%, similar to some of the previous studies on BSI caused by K. pneumoniae [7]–[10]. Analyzing phylogroups in relation to mortality, KpIII had the highest 30-day mortality (10/34, 29.4%). To the best to our knowledge, this is the first report indicating a relatively high mortality associated with K. variicola. The difference in mortality between KpI and KpIII was significant (P = 0.037) both in univariate analysis and in multivariate analysis adjusting for age, comorbidity, gender, time to adequate antimicrobial therapy, and polymicrobial infections. The difference in mortality between the clades cannot be explained by differences in clinical characteristics of patients since they were similar (Table 4). Measuring time to adequate antimicrobial treatment and other supportive therapy is complicated in this type of study as severity of disease and presentation of disease often influences choice and timing of empirical treatment. However as resistance rates were low in our study more than half of the patients (55.4%) received adequate antibiotic treatment within 4 hours. After 24 hours most patients (87.1%) had received adequate antibiotic treatment, and there were no differences among the survivors and the deaths in time to adequate antibiotics.
However there are limitations to consider. The major concern is the small number of events (30-d mortality of 23 patients in the two analyzed phylogroups), causing limitations in the multivariate model. Despite this concern, it is interesting that our data suggest that the underestimated pathogen K. variicola is associated with poor prognosis in BSI. The 30-d mortality is often used as an endpoint in studies measuring mortality. However, when studying BSI and mortality, there is always a risk that the cause of death is not attributable to the BSI, although this risk would be regarded equally large in all phylogroups. It could be debated whether other variables could be adjusted for, but the risk of this strategy is that such variables could be on the causal pathway between phylogroup and mortality.
Numerous hospital-based studies have notified that several comorbidities such as malignancy, diabetes mellitus, cirrhosis, biliary tract disorders and alcoholism may impair patient defenses and hence contribute to the development of K. pneumoniae infection [1],[7],[9],[70]–[73]. Previous studies on BSI caused by K. pneumoniae have shown that the disease often affects older patients with high comorbidity. Our study corroborates these earlier findings. Most of the patients in our study were male with a median age of 70, 32.4% had Charlson comorbidity index 4 or more, and almost half of the episodes were hospital acquired. Increasing Charlson comorbidity index was strongly associated with mortality (P = 0.005). Advanced age was associated with an increased, but not significant, risk of death. There was a similar amount of hospital-acquired infections among the deaths and survivors.
In accordance to what was reported previously [37],[46], our study demonstrates that isolates belonging to the phylogroup KpI were found to be more resistant than K. variicola isolates. From the present study, we retrieved only five MDR isolates that were ESBL-producers and belonged to KpI. Recently, Valverde et al. [74] determined the phylogenetic structure of a collection of ESBL-producing K. pneumoniae isolates. Expectedly, most of the ESBL-producing isolates belonged to KpI. A minority of isolates was classified in the KpIII group, and almost all of them were CTX-M-10 producers. Lastly, the prevalence of class 1 integrons was screened among these three phylogroups; the class 1 integrons were dominantly associated with KpI but very few were observed in isolates of KpIII [75]. As suggested previously, the high prevalence of KpI isolates among clinical isolates could be a reflection of the higher antimicrobial resistance rates within this group.
This collection of K. pneumoniae isolates contained few virulence factors compared with reports from previous studies, based on community-acquired BSI and severe disease. Mucoid phenotype (n = 8), virulence genes rmpA, wcaG and allS (n = 21 isolates; two of them had both wcaG and allS), and capsular serotypes K1, K2, K5, K20, K54 and K57 (n = 18) were all relatively rare in the collection. Much effort has been made in the last decades to estimate the link between the capsular types, virulence potential, and the nature of diseases, but mostly with community-acquired isolates, which differ in ST and virulence factors from nosocomial isolates [6],[21],[22],[31],[76]. Within the phyologroups in our dataset, there was no correlation between the serotypes or virulence genes and CCs or STs.
Previous reports have expressed concern about numerous bacterial virulence genes which were significantly associated with invasive strains. Herein, we show that rmpA gene was associated only to K2, K20 and K57, and not to K1, K5 and K54 as found in previous reports [51],[77]. In the same reports wcaG, encoding capsular fucose production, was associated with capsular types K1 and K54 [51],[77], which was further corroborated by our present results. Although the presence of the wcaG gene has not been well studied, there is data to suggest that strains harbouring this gene are more often observed in severe disease [51]. Interestingly, two fatal strains harboring the wcaG gene were recorded in our study (Table S1); ST616 (K54, wcaG, KpIII) and ST604 (wcaG/allS, KpII). Conversely, the six isolates harboring the allS gene did not belong to any of the investigated capsular types. Noteworthy, this gene was not universally associated to K1 serotype, but considered as a specific marker for the hypervirulent clone (CC23 K1) isolated from patients with pyogenic liver abscesses detected in several countries [21],[34],[47],[51],[76],[78]–[80]. In a recent report from France, five cases of fatal BSI were attributed to K. pneumoniae serotype K2, possessing the rmpA gene [47]. The isolates belonged to two clones; ST86 (two isolates) and ST380 (3 isolates). The same genotype (ST380, K2, rmpA) was encountered in our study but was not associated with a fatal outcome. In the 30-day mortality group (24/139 patients) only three isolates featured one or more virulence factors. In this study, where both community- and hospital-acquired BSIs were included, no indication for an existing correlation between above mentioned virulence factors and increased risk of mortality in BSI caused by K. pneumoniae could be found.
Conclusions
We have analyzed the population structure of invasive clinical isolates of K. pneumonia sensu lato in Sweden, and found K. pneumoniae (KpI), but also K. quasipneumoniae (KpII) and K. variicola (KpIII). The 30-d mortality was more commonly associated with infecting strains belonging to K. variicola, but was not associated with known virulence factors, including some of the most virulent capsular types. More studies with enhanced species identification would be valuable to expand the data on the clinical importance of K. variicola in relation to K. pneumoniae sensu stricto. In general, a high level of comorbidity, equally high among the patients with BSI caused by K. pneumoniae as among the patients with BSI caused by K. variicola, was observed.
Supporting Information
Circular Neighbor-joining tree of 139 isolates based on the concatenated sequences of seven MLST loci based on a Jukes-Cantor distance matrix. Main bootstrap values obtained are highlighted on at the main node of the phylogeny. The tree was rooted using the nucleotide sequences of the seven genes of E. coli and considered as an out-group. The tree displays all the STs into their phylogroups. Colors of isolates symbols are specific of each phylogroups. Blue color corresponds to the KpI phylogroup, green corresponds to KpII, and red corresponds to KpIII. Empty symbols correspond to the references strains specific for each phylogroups.
(TIF)
Minimal Spanning Trees (MSTs) analysis of Klebsiella pneumoniae strains based on MLST allelic profiles. Each circle corresponds to an ST. The area of each circle corresponds to the number of isolates. The relationships between strains are indicated by the connections between the isolates and the lengths of the branches linking them. Black lines connecting pairs of STs indicate that they differ in one allele (thick lines), two and three alleles (thin), or four to seven alleles (dashed). Four MST graphs were generated separately based on the following associations. A: MST vs mortality, B: MST vs serotypes, C: MST vs virulence genes and D: MST vs mucoid phenotype.
(TIF)
Database displaying the genotypic and phenotypic features of the studied strains. Strains ID, MLST STs, Allelic profiles, phylogroups, patient sex, patient age, death within 30 days: 1 = yes, malignancy, Charlson comorbidity index, nosocomial (>48 h after admittance or < = 30 days after discharge), time to adequate antimicrobial therapy, mucoid phenotype, serotype and virulence factors.
(XLSX)
Acknowledgments
We thank biomedical scientists Ms. Marie Andersson and Mr. Patrik Jonsson at the Department of Clinical microbiology, Karolinska University Hospital, for excellent technical assistance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a grant from The European Society for Clinical Microbiology and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. . Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, et al. (1998) Primary liver abscess due to Klebsiella pneumoniae in Taiwan. . Clin Infect Dis 26:1434–1438. [DOI] [PubMed] [Google Scholar]
- 3. Rahimian J, Wilson T, Oram V, Holzman RS (2004) Pyogenic liver abscess: recent trends in etiology and mortality. . Clin Infect Dis 39:1654–1659. [DOI] [PubMed] [Google Scholar]
- 4. Lu CH, Chang WN, Chang HW (2002) Klebsiella meningitis in adults: clinical features, prognostic factors and therapeutic outcomes. . Journal of Clinical Neuroscience 9:533–538. [DOI] [PubMed] [Google Scholar]
- 5. Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, et al. (2002) Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. . Emerg Infect Dis 8:160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, et al. (2007) Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. . Emerg Infect Dis 13:986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB (2009) Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. . Am J Med 122:866–873. [DOI] [PubMed] [Google Scholar]
- 8. Thom KA, Schweizer ML, Osih RB, McGregor JC, Furuno JP, et al. (2008) Impact of empiric antimicrobial therapy on outcomes in patients with Escherichia coli and Klebsiella pneumoniae bacteremia: a cohort study. . BMC Infect Dis 8:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsay RW, Siu LK, Fung CP, Chang FY (2002) Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. . Arch Intern Med 162:1021–1027. [DOI] [PubMed] [Google Scholar]
- 10. Jung Y, Lee MJ, Sin HY, Kim NH, Hwang JH, et al. (2012) Differences in characteristics between healthcare-associated and community-acquired infection in community-onset Klebsiella pneumoniae bloodstream infection in Korea. . BMC Infect Dis 12:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donnarumma F, Indorato C, Mastromei G, Goti E, Nicoletti P, et al. (2011) Molecular analysis of population structure and antibiotic resistance of Klebsiella isolates from a three-year surveillance program in Florence hospitals, Italy. . Eur J Clin Microbiol Infect Dis 31:371–378. [DOI] [PubMed] [Google Scholar]
- 12. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, et al. (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. . Clin Infect Dis 48:1–12. [DOI] [PubMed] [Google Scholar]
- 13. Livermore DM (2009) Has the era of untreatable infections arrived? J Antimicrob Chemother 64 Suppl. 1: i29–36. [DOI] [PubMed] [Google Scholar]
- 14. Cagnacci S, Gualco L, Roveta S, Mannelli S, Borgianni L, et al. (2008) Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-beta-lactamase: first Italian outbreak. . J Antimicrob Chemother 61:296–300. [DOI] [PubMed] [Google Scholar]
- 15.Vatopoulos A (2008) High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece-a review of the current evidence. Euro Surveill. 13. [PubMed]
- 16. Peirano G, Sang JH, Pitondo-Silva A, Laupland KB, Pitout JD (2012) Molecular epidemiology of extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. . J Antimicrob Chemother 67:1114–1120. [DOI] [PubMed] [Google Scholar]
- 17. Warburg G, Hidalgo-Grass C, Partridge SR, Tolmasky ME, Temper V, et al. (2012) A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. . J Antimicrob Chemother 67:898–901. [DOI] [PubMed] [Google Scholar]
- 18. Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, et al. (2009) Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. . J Antimicrob Chemother 63:654–658. [DOI] [PubMed] [Google Scholar]
- 19. Yeh KM, Lin JC, Yin FY, Fung CP, Hung HC, et al. (2010) Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. . J Infect Dis 201:1259–1267. [DOI] [PubMed] [Google Scholar]
- 20. Podschun R, Pietsch S, Holler C, Ullmann U (2001) Incidence of Klebsiella species in surface waters and their expression of virulence factors. . Appl Environ Microbiol 67:3325–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, et al. (2009) Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. . PLoS One 4:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shon AS, Russo TA (2012) Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 7:669–671. [DOI] [PubMed] [Google Scholar]
- 23. Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, et al. (2008) Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. . J Clin Microbiol 46:2231–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orskov I, Orskov F (1984) Serotyping of Klebsiella: Academic Press.
- 25. Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, et al. (2007) Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. . Clin Infect Dis 45:284–293. [DOI] [PubMed] [Google Scholar]
- 26. Cortes G, Borrell N, de Astorza B, Gomez C, Sauleda J, et al. (2002) Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. . Infect Immun 70:2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomas JM, Camprubi S, Merino S, Davey MR, Williams P (1991) Surface exposure of O1 serotype lipopolysaccharide in Klebsiella pneumoniae strains expressing different K antigens. . Infect Immun 59:2006–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A (2003) Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. . Res Microbiol 154:9–16. [DOI] [PubMed] [Google Scholar]
- 29. Struve C, Bojer M, Krogfelt KA (2008) Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. . Infect Immun 76:4055–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin WH, Wang MC, Tseng CC, Ko WC, Wu AB, et al. (2010) Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. . Infection 38:459–464. [DOI] [PubMed] [Google Scholar]
- 31. Shon AS, Bajwa RP, Russo TA (2013) Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. . Virulence 4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT (2011) The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. . Microbiology 157:3446–3457. [DOI] [PubMed] [Google Scholar]
- 33. Wu JH, Wu AM, Tsai CG, Chang XY, Tsai SF, et al. (2008) Contribution of fucose-containing capsules in Klebsiella pneumoniae to bacterial virulence in mice. . Exp Biol Med (Maywood) 233:64–70. [DOI] [PubMed] [Google Scholar]
- 34. Chou HC, Lee CZ, Ma LC, Fang CT, Chang SC, et al. (2004) Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. . Infect Immun 72:3783–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brisse S GF, Grimont PAD (2006) The genus Klebsiella In: Dworkin M FS, Rosenberg E, Schleifer K-H, Stackebrandt E, editor. The Prokaryotes • A Handbook on the Biology of Bacteria. 3rd edition ed. New York: Springer.
- 36. Brisse S, Verhoef J (2001) Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. . Int J Syst Evol Microbiol 51:915–924. [DOI] [PubMed] [Google Scholar]
- 37. Brisse S, van Himbergen T, Kusters K, Verhoef J (2004) Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. . Clin Microbiol Infect 10:942–945. [DOI] [PubMed] [Google Scholar]
- 38. Haeggman S, Lofdahl S, Paauw A, Verhoef J, Brisse S (2004) Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. . Antimicrob Agents Chemother 48:2400–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fevre C, Passet V, Weill FX, Grimont PA, Brisse S (2005) Variants of the Klebsiella pneumoniae OKP chromosomal beta-lactamase are divided into two main groups, OKP-A and OKP-B. . Antimicrob Agents Chemother 49:5149–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang M, Cao B, Yu Q, Liu L, Gao Q, et al. (2008) Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species. . J. Clin Microbiol 46:3555–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fevre C, Passet V, Deletoile A, Barbe V, Frangeul L, et al. (2011) PCR-based identification of Klebsiella pneumoniae subsp. rhinoscleromatis, the agent of rhinoscleroma. . PLoS Negl Trop Dis 5:e1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenblueth M, Martinez L, Silva J, Martinez-Romero E (2004) Klebsiella variicola, a novel species with clinical and plant-associated isolates. . Syst Appl Microbiol 27:27–35. [DOI] [PubMed] [Google Scholar]
- 43.Brisse S, Passet V, Grimont PA (2014) Description of Klebsiella quasipneumoniae sp. nov., a novel species isolated from human infections, with two subspecies Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that K. singaporensis is a junior heterotypic synonym of K. variicola. Int J. Syst Evol Microbiol. [DOI] [PubMed]
- 44.Grimont PAD GF (2005) Genus Klebsiella. In:Brenner DJ, Krieg NR SJeditors. Bergey’s manual of Systematic Bacteriology. New York: Springer-Verlag 685–693.
- 45. Alves MS, Dias RC, de Castro AC, Riley LW, Moreira BM (2006) Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. . J. Clin Microbiol 44:3640–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Melo ME, Cabral AB, Maciel MA, da Silveira VM, de Souza Lopes AC (2011) Phylogenetic groups among Klebsiella pneumoniae isolates from Brazil: relationship with antimicrobial resistance and origin. . Curr Microbiol 62:1596–1601. [DOI] [PubMed] [Google Scholar]
- 47. Decre D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, et al. (2011) Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. . J Clin Microbiol 49:3012–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng HP, Chang FY, Fung CP, Siu LK (2002) Klebsiella pneumoniae liver abscess in Taiwan is not caused by a clonal spread strain. . J Microbiol Immunol Infect 35:85–88. [PubMed] [Google Scholar]
- 49. Ma LC, Fang CT, Lee CZ, Shun CT, Wang JT (2005) Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. . J Infect Dis 192:117–128. [DOI] [PubMed] [Google Scholar]
- 50.Olsson-Liljequist B, Larsson P, Walder M, Miorner H (1997) Antimicrobial susceptibility testing in Sweden. III. Methodology for susceptibility testing. Scand J Infect Dis Suppl. 10513–23. [PubMed]
- 51. Turton JF, Perry C, Elgohari S, Hampton CV (2010) PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. . J Med Microbiol 59:541–547. [DOI] [PubMed] [Google Scholar]
- 52. Brisse S, Issenhuth-Jeanjean S, Grimont PA (2004) Molecular serotyping of Klebsiella species isolates by restriction of the amplified capsular antigen gene cluster. . J Clin Microbiol 42:3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giske CG, Froding I, Hasan CM, Turlej-Rogacka A, Toleman M, et al. (2012) Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. . Antimicrob Agents Chemother 56:2735–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. . J Clin Microbiol 43:4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. . Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. . Mol Biol Evol 21:255–265. [DOI] [PubMed] [Google Scholar]
- 57. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. . Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- 58. Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, et al. (2011) Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. . PLoS One 6:e25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. . J Bacteriol 186:1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Charlson ME, Pompei P, Ales KL, MacKenzie CR (1987) A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. . J Chronic Dis 40:373–383. [DOI] [PubMed] [Google Scholar]
- 61. Kjellander C, Bjorkholm M, Cherif H, Kalin M, Giske CG (2012) Hematological: Low all-cause mortality and low occurrence of antimicrobial resistance in hematological patients with bacteremia receiving no antibacterial prophylaxis: a single-center study. . Eur J Haematol 88:422–430. [DOI] [PubMed] [Google Scholar]
- 62.Hasan CM, Turlej-Rogacka A, Vatopoulos AC, Giakkoupi P, Maatallah M, et al. (2013) Dissemination of bla in Greece at the peak of the epidemic of 2005–2006: clonal expansion of Klebsiella pneumoniae clonal complex 147. Clin Microbiol Infect. [DOI] [PubMed]
- 63. Breurec S, Guessennd N, Timinouni M, Le TA, Cao V, et al. (2013) Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. . Clin Microbiol Infect 19:349–355. [DOI] [PubMed] [Google Scholar]
- 64. Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, et al. (2006) First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. . Antimicrob Agents Chemother 50:2880–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, et al. (2010) Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. . Emerg Infect Dis 16:1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Suh JY, Son JS, Chung DR, Peck KR, Ko KS, et al. (2010) Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. . Antimicrob Agents Chemother 54:560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. . Clin Microbiol Infect 18:268–281. [DOI] [PubMed] [Google Scholar]
- 68. Samuelsen O, Toleman MA, Hasseltvedt V, Fuursted K, Leegaard TM, et al. (2011) Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. . Clin Microbiol Infect 17:1811–1816. [DOI] [PubMed] [Google Scholar]
- 69. Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, et al. (2013) Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. . J Med Microbiol 62:801–803. [DOI] [PubMed] [Google Scholar]
- 70. Wang LS, Lee FY, Cheng DL, Liu CY, Hinthorn DR, et al. (1990) Klebsiella pneumoniae bacteremia: analysis of 100 episodes. . J Formos Med Assoc 89:756–763. [PubMed] [Google Scholar]
- 71. Tsai SS, Huang JC, Chen ST, Sun JH, Wang CC, et al. (2010) Characteristics of Klebsiella pneumoniae bacteremia in community-acquired and nosocomial infections in diabetic patients. . Chang Gung Med J 33:532–539. [PubMed] [Google Scholar]
- 72. Lee KH, Hui KP, Tan WC, Lim TK (1994) Klebsiella bacteraemia: a report of 101 cases from National University Hospital, Singapore. . J Hosp Infect 27:299–305. [DOI] [PubMed] [Google Scholar]
- 73. Skogberg K, Lyytikainen O, Ruutu P, Ollgren J, Nuorti JP (2008) Increase in bloodstream infections in Finland, 1995–2002. . Epidemiol Infect 136:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Valverde A, Coque TM, Garcia-San Miguel L, Baquero F, Canton R (2008) Complex molecular epidemiology of extended-spectrum beta-lactamases in Klebsiella pneumoniae: a long-term perspective from a single institution in Madrid. . J Antimicrob Chemother 61:64–72. [DOI] [PubMed] [Google Scholar]
- 75. Lima AM, Melo ME, Alves LC, Brayner FA, Lopes AC (2014) Investigation of class 1 integrons in Klebsiella pneumoniae clinical and microbiota isolates belonging to different phylogenetic groups in Recife, State of Pernambuco. . Rev Soc Bras Med Trop 47:165–169. [DOI] [PubMed] [Google Scholar]
- 76. Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, et al. (2007) Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. . J Med Microbiol 56:593–597. [DOI] [PubMed] [Google Scholar]
- 77. Chen Z, Liu M, Cui Y, Wang L, Zhang Y, et al. (2014) A novel PCR-based genotyping scheme for clinical Klebsiella pneumoniae. . Future Microbiol 9:21–32. [DOI] [PubMed] [Google Scholar]
- 78. Merlet A, Cazanave C, Dutronc H, de Barbeyrac B, Brisse S, et al. (2012) Primary liver abscess due to CC23-K1 virulent clone of Klebsiella pneumoniae in France. . Clin Microbiol Infect 18:E338–339. [DOI] [PubMed] [Google Scholar]
- 79. Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, et al. (2011) Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. . Antimicrob Agents Chemother 55:5493–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gundestrup S, Struve C, Stahlhut SG, Hansen DS (2014) First Case of Liver Abscess in Scandinavia Due to the International Hypervirulent Klebsiella Pneumoniae Clone ST23. Open Microbiol J. 822–24. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circular Neighbor-joining tree of 139 isolates based on the concatenated sequences of seven MLST loci based on a Jukes-Cantor distance matrix. Main bootstrap values obtained are highlighted on at the main node of the phylogeny. The tree was rooted using the nucleotide sequences of the seven genes of E. coli and considered as an out-group. The tree displays all the STs into their phylogroups. Colors of isolates symbols are specific of each phylogroups. Blue color corresponds to the KpI phylogroup, green corresponds to KpII, and red corresponds to KpIII. Empty symbols correspond to the references strains specific for each phylogroups.
(TIF)
Minimal Spanning Trees (MSTs) analysis of Klebsiella pneumoniae strains based on MLST allelic profiles. Each circle corresponds to an ST. The area of each circle corresponds to the number of isolates. The relationships between strains are indicated by the connections between the isolates and the lengths of the branches linking them. Black lines connecting pairs of STs indicate that they differ in one allele (thick lines), two and three alleles (thin), or four to seven alleles (dashed). Four MST graphs were generated separately based on the following associations. A: MST vs mortality, B: MST vs serotypes, C: MST vs virulence genes and D: MST vs mucoid phenotype.
(TIF)
Database displaying the genotypic and phenotypic features of the studied strains. Strains ID, MLST STs, Allelic profiles, phylogroups, patient sex, patient age, death within 30 days: 1 = yes, malignancy, Charlson comorbidity index, nosocomial (>48 h after admittance or < = 30 days after discharge), time to adequate antimicrobial therapy, mucoid phenotype, serotype and virulence factors.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.