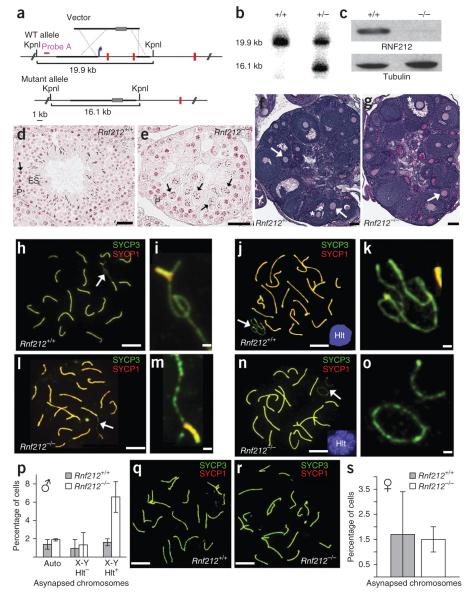
Figure 3.
Gonad morphology and homolog synapsis in Rnf212−/− knockout mice. (a) Rnf212 targeting scheme. Thick lines represent homology arms used for targeting. Gray rectangles represent the neomycin-resistance cassette. The blue arrow indicates the position of the promoter, and red vertical lines represent Rnf212 exons (not to scale). WT, wild type. (b) Southern analysis of KpnI-digested genomic DNA from wild-type and heterozygous embryonic stem cells hybridized with the probe shown in a. (c) Protein blot analysis of RNF212 in protein extracts from whole testes. (d,e) Representative seminiferous tubules stained with hematoxylin and eosin from testis sections from wild-type (d) and Rnf212−/− (e) mice. P, pachytene-stage cells; ES, elongated spermatids. Arrows indicate metaphase I cells. In Rnf212−/− tubules, cells progress to metaphase I (stage XII), but normal chromosome congression is not observed, and postmeiotic spermatogenic cells (spermatids and spermatozoa) are absent. (f,g) Representative ovary sections stained with periodic acid Schiff and hematoxylin from wild-type (f) and Rnf212−/− (g) mice. Asterisks indicate antral follicles with defined antral space. Arrows highlight secondary follicles surrounded by more than one layer of cuboidal granulosa cells. (h–o) Spread spermatocyte nuclei immunostained for SYCP3 and SYCP1. (h,i) Early pachynema nucleus from wild-type mouse (h), with magnified view of the sex chromosomes indicated by the arrow (i). (j,k) Midpachynema nucleus from wild-type cells (j). The inset shows H1t staining. (k) Magnified view of the indicated sex chromosomes in j. (l,m) Early pachynema nucleus from Rnf212−/− cells (l), with magnified view of the sex chromosomes indicated by an arrow (m). (n,o) Midpachynema nucleus from Rnf212−/− cells (n), with magnified view of the sex chromosomes (o). (p) Quantification of synapsis defects in pachytene spermatocytes. Numbers of nuclei analyzed in wild-type and Rnf212−/− cells, respectively: 293 and 521 for autosomal asynapsis (auto) and X-Y asynapsis in H1t-negative pachytene cells; 686 and 622 for X-Y asynapsis in H1t-positive pachytene cells. (q,r) Pachytene-stage fetal oocytes from wild-type (q) and Rnf212−/− mutant (r) animals immunostained for SYCP3 and SYCP1. (s) Quantification of synapsis defects in pachytene oocytes (191 and 272 wild-type and Rnf212−/− nuclei, respectively). Error bars in p and s, s.e.m. Scale bars, 100 μm in d–g, 10 μm in h,j,l,n,q,r and 1 μm in i,k,m,o.