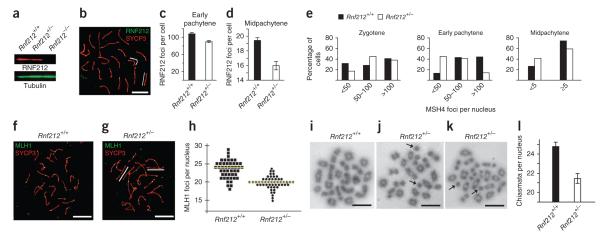
Figure 6.
Rnf212 is haploinsufficient. (a) Quantitative protein blot analysis of RNF212 in testis cell extracts from wild-type, Rnf212+/− and Rnf212−/− mice. (b) Midpachytene Rnf212+/− spermatocyte nucleus immunostained for RNF212 and SYCP3. White lines highlight synaptonemal complexes that lack RNF212 foci. (c,d) Numbers of RNF212 foci per nucleus ± s.e.m. in wild-type and Rnf212+/− spermatocytes at early pachynema (c) and midpachynema (d). (e) Numbers of MSH4 foci per nucleus in wild-type and Rnf212+/− spermatocytes at successive prophase substages. Statistical comparisons: zygotene, P = 0.04 (G test; n = 83 Rnf212+/+ and 68 Rnf212+/− nuclei); early pachytene, P = 9 × 10−13 (G test; n = 131 Rnf212+/+ and 270 Rnf212−/− nuclei); midpachytene, P = 0.01 χ-square test; n = 121 Rnf212+/+ and 126 Rnf212−/− nuclei). (f,g) Midpachytene spermatocyte nuclei from wild-type (f) and Rnf212+/− (g) mice immunolabeled for MLH1 and SYCP3. White lines in g highlight synaptonemal complexes that lack MLH1 foci. (h) Numbers of MLH1 foci per nucleus in midpachytene-stage spermatocytes. Each symbol represents a single nucleus. Yellow lines indicate the average numbers of foci. (i–k) Chromosome spreads of spermatocytes in diakinesis/metaphase I from Rnf212+/+ (i) and Rnf212+/− (j,k) mice. Independent examples from Rnf212−/− spermatocytes are shown in (j) and (k). Arrows in j and k highlight achiasmate univalent chromosomes. (l) Numbers of chiasmata per nucleus (± s.e.m.) in wild-type and Rnf212+/− diakinesis/metaphase I spermatocytes. Scale bars, 10 μm.