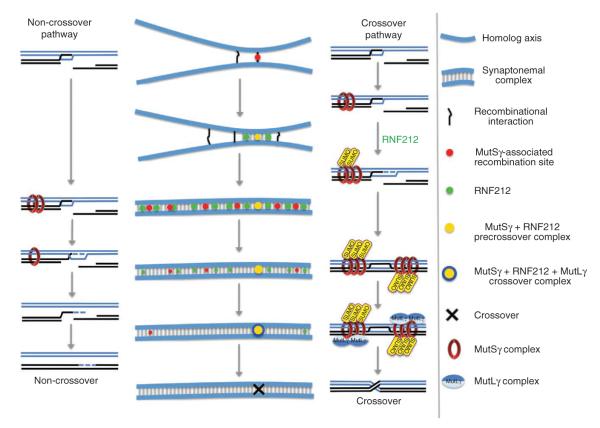
Figure 7.
Summary and model of RNF212 function. Schematics showing the cytological development of recombination complexes and parallel molecular pathways of crossover and non-crossover recombination. Black and blue lines represent homologous DNA duplexes. Although four chromatids are present at this stage, for simplicity, only the two chromatids involved in recombination are shown. Both crossover and non-crossover pathways initiate from a common D-loop precursor. As synapsis ensues, binding of the MutSγ complex initially stabilizes most or all D-loops. In the absence of RNF212-mediated stabilization, MutSγ dissociates, and D-loops are unwound, resulting in non-crossover formation. At crossover sites, RNF212-dependent SUMOylation enhances the association of MutSγ, D-loops are stabilized, and formation of crossover-specific double Holliday junctions ensues. These crossover precursors become competent to assemble the crossover-specific resolution factor, MutLγ, and crossing-over occurs. Recombination sites that nucleate synaptonemal complex formation have a high probability of being the first sites where MutSγ and RNF212 colocalize. At such sites, a positive feedback loop locally enhances the binding of both MutSγ and RNF212. General binding of RNF212 to sites along the synaptonemal complex central element disfavors stabilization of MutSγ at other recombination sites.