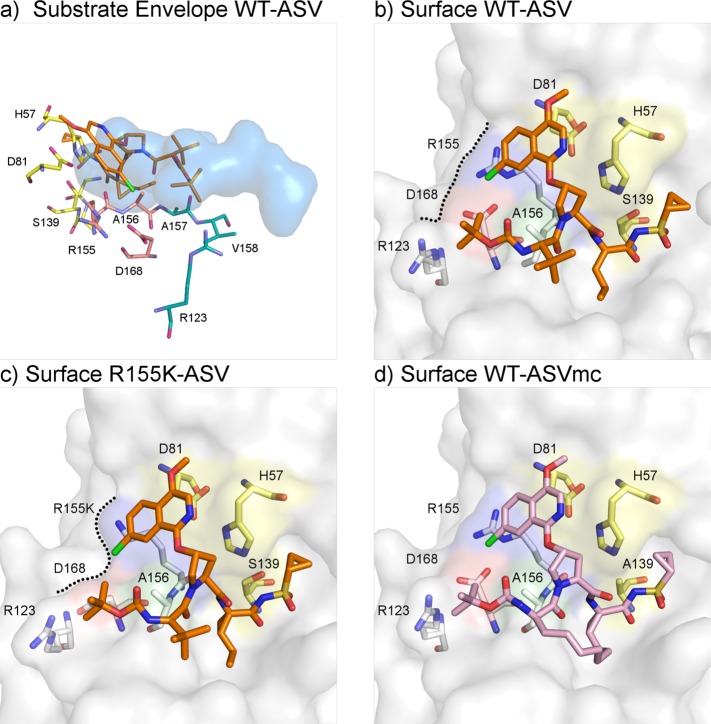
Figure 1.
Asunaprevir’s binding mode is reliant on protease S2 residues: structure of HCV NS3/4A protease in complex with ASV. (a) ASV P2* isoquinoline protrudes from the substrate envelope (blue volume), interacting with protease S2 residues Arg155 and Asp168 (orange). (b) In the WT complex (PDB id: 4WF8), ASV (orange sticks) engages in stacking interactions with the catalytic D81 and S2 residue R155. (c) In the R155K complex (PDB id: 4WH6), Lys155 is no longer stabilized by Asp168, which rotates toward R123 for electrostatic interaction. Consequently, ASV’s P2* isoquinoline loses important interaction binding surface (black dashed lines) and is destabilized. (d) ASVmc (PDB id: 4WH8, pink sticks) adopts a similar binding mode as ASV.