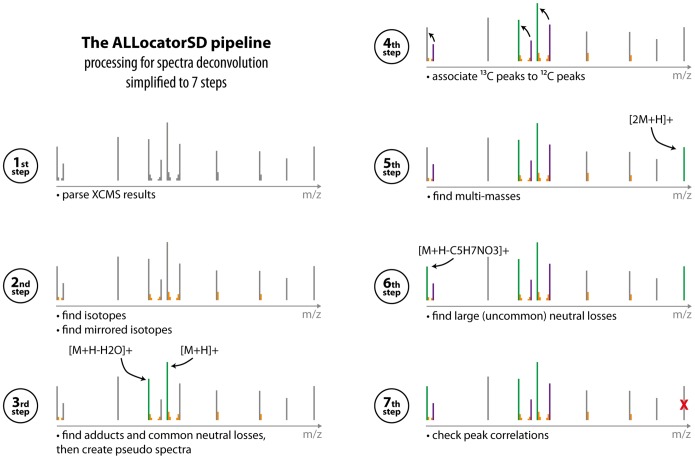
Figure 2. Summary of the ALLocatorSD pipeline for pseudo spectra allocation and peak annotation, divided into seven steps:
Reading peaks from the XCMS output (1st step), identification of isotopes (2nd step), computation of pseudo spectra based on common adducts and neutral losses (3rd step), association of 13C monoisotopic peaks to 12C monoisotopic peaks (4th step), identification of homoadducts (5th step), identification of large (uncommon) neutral losses (6th step), and a check for peak correlations to validate pseudo spectra (7th step).